抽象的
背景
癌症的早期发现和诊断对于改善患者的预后至关重要。人工智能(AI)模型在癌症的早期检测和诊断中表现出了希望,但是关于完全利用电子健康记录中存储的纵向数据的方法的证据有限。这篇综述旨在总结目前用于从纵向数据预测癌症的方法,并提供有关如何开发此类模型的建议。
方法
审查是在Prisma-SCR指导下进行的。搜索了六个数据库(Medline,Embase,Web of Science,IEEE Xplore,PubMed和Scopus),以搜索2/2/2024之前发布的相关记录。搜索术语与人工智能,预测,健康记录,纵向和癌症有关。提取了与文章的几个领域有关的数据:(1)出版细节,(2)研究特征,(3)输入数据,(4)模型特征,(4)可重复性,(5)使用Probast工具进行质量评估。根据与癌症检测和风险预测模型报告有关的术语框架进行了评估。
结果
在筛选的653个记录中,审查中包括33个记录;10预测癌症的风险,18例进行癌症检测或早期检测,4例预测复发,1例预测转移。研究中预测的最常见的癌症是结直肠癌(n= 9)和胰腺癌(n= 9)。16研究使用功能工程来表示时间数据,其中最常见的特征代表趋势。18使用了深度学习模型,这些模型采用了直接的顺序输入,最常见的神经网络,还包括卷积神经网络和变压器。在研究之间,预测窗口和提前时间也有很大的不同,即使对于预测相同癌症的模型也是如此。在90%的研究中发现了高偏见的高风险。由于研究设计不当,通常引入这种风险(n= 26)和样本量(n= 26)。结论这篇评论强调了纵向数据的癌症预测方法的广度。
我们确定了可以改善方法报告的领域,特别是在患者轨迹中应用模型的何处。
该评论显示了进一步工作的机会,包括对这些方法的比较及其在其他癌症中的应用。
背景
癌症是全球死亡的主要原因,癌症的负担每年持续增长,在2022年报告约有2000万例和1000万例死亡,预计2050年将达到3500万[1]。癌症的早期诊断可能会导致由于较早的干预和可能增加的治疗选择范围而改善结果[2,,,,3]。预测模型旨在通过发现患癌症的高风险,对癌症进行早期检测或通过向转移或复发风险的患者发出信号来促进早期诊断[4]。这些模型有可能影响未来的研究,或者通过用作决策支持工具直接影响护理。传统上,使用统计方法构建了风险预测模型,但是,人工智能(AI)表明了有望提高模型的预测能力[5]。AI是一组旨在模仿人类决策的技术[6]。更具体地说,AI技术是开发的系统,可以根据人类目标提供信息的输出[7]。机器学习(ML)是该领域的一个子集,它利用从经验中学习的算法来优化定义的任务。尽管这些术语具有不同的含义,但在该字段中,鉴于最适合本应用程序的AI子集,它们通常会互换使用。
癌症预测模型经常使用电子健康记录(EHR),该记录包含医疗保健专业人员在患者护理过程中收集的回顾性数据,例如,实验室检查或诊断。这些记录包含常规收集的患者数据,用于告知预测。EHR的使用和可用性越来越多地为预测模型提供了大量数据,并允许以传统前瞻性研究成本的一小部分进行回顾性研究[8]。
许多癌症预测模型使用横截面方法使用EHR数据,而无需考虑数据的时间方面。但是,可以探索纵向数据以充分利用EHRS中存储的信息[8]。应从上下文中查看患者的测量 - 随着时间的推移,随着时间的变化可能会提供有关患者健康的更多信息,而不是观察静态观察,并且最新的观察结果可能比遥远的观察更能提供信息。例如,虽然长期存在的糖尿病是胰腺癌的危险因素,但已建议新发糖尿病是无症状癌症的指标[9,,,,10,,,,11]。诸如实验室测试之类的数量也会受到患者间的可变性,这些值的变化可能比瞬时测量更具信息性[12]。纵向数据已成功地用于其他医疗领域,例如死亡率和败血症预测[13,,,,14,,,,15]。
需要分析纵向数据的问题发生在许多应用中,例如气象,金融,运输和音频处理[16,,,,17]。由于时间性引入的特定挑战,这些方案需要专门的方法,包括连续输入与高维度之间的相关性,在该方面将时间点视为单个输入。医疗保健中的时间序列数据提出了特定的其他挑战,包括:数据不规则性,因为观察之间的时间间隔通常是不规则的;数据稀疏性是由于不频繁的医疗相互作用和分类医学遭遇的一式式编码而导致的;数据异质性,指的是高度多样化的轨迹和患者的结果;和模型不透明度,因为时间序列建模的许多模型都需要不可解释的复杂方法[18]。
许多评论已经评估了用于纵向健康数据的方法[18,,,,19,,,,20]。Cascarano等。提供了可能应用于纵向生物医学数据的ML方法的叙述性回顾[20],而Carrasco-Ribelles等。系统地审查了使用EHR的纵向数据进行基于AI的预测模型的研究[19]和Xie等。系统地回顾了代表健康记录中时间数据的深度学习方法。没有一个专注于癌症并纳入所有可能的方法,包括深度学习和特征工程方法。在当前的评论中,保持对癌症预测的关注意味着审查的研究正在解决共同的数据挑战。由于癌症的长期和渐进性,它们通常会依靠较长时间内收集的稀疏数据。
此外,确定的最新系统评论已发布到2022年1月的记录[19]。在这样一个快速移动的领域中,可能在当前工作之前的两年中开发了使用的方法。卡斯卡拉诺(Cascarano20]。为了充分实现癌症预测模型在改善全球癌症结果方面的潜力,预测模型在开发和验证中都需要严格的方法。迄今为止,尚无评论检查研究纵向数据进行癌症预测的研究质量。
这篇综述旨在提供用于通过EHR中纵向数据预测癌症的AI方法的摘要。
为此,进行了范围审查[21]。评论的目标如下:
-
1。
识别并总结用于从EHR中存储的纵向数据预测癌症的方法。
-
2。
评估预测模型中使用的时间窗口针对框架。
-
3。
确定癌症预测研究中增加偏见风险的共同领域,以使用偏见评估工具的预测模型风险(probast)指导未来的研究[22]。
-
4。
为癌症的纵向预测模型设计和报告提供了建议。
方法
这项范围审查遵循首选的报告项目,用于系统审查和荟萃分析的扩展,用于范围审查(Prisma-SCR)[23]。
数据库和搜索策略
搜索策略是由作者迭代制定的。搜索了六个数据库:Medline,Embase,Science Web,IEEE Xplore,Scopus和PubMed。搜索策略适用于每个数据库,但是,每个搜索都包含了与范围审查问题相关的每个概念有关的术语。这些概念是人工智能,“预测”,“ Ehrs”,“纵向”和“癌症”。完整的搜索词在附加文件1中提供。搜索是在15上进行的Th2023年8月,两者都更新了2ND2024年2月和9Th2024年8月,一年没有限制。搜索了引用和参考列表,以获取每项合格的研究,以检索初始搜索中未检索的其他记录。
资格标准和研究选择
所有记录均进口给Rayyan,这是一个为支持系统评价的平台而开发的,并删除了重复项。
然后通过V.M筛选标题和摘要,并根据排除标准进行评估。在纳入标准内有歧义的地方,与L.S.讨论了记录。和O.J.直到做出决定。将结果记录筛选为全文资格。
符合条件的研究是使用来自EHR的纵向数据来预测癌症的研究,包括转移和复发的预测。EHR数据不仅限于结构化数据。
根据以下标准排除文章:
-
1。
不是主要的研究文章。
-
2。
该研究不能预测癌症。
-
3。
该研究不使用AI/ML方法。
-
4。
该研究不使用纵向预测因子。
-
5。
该研究纯粹是一项实施或验证研究。
-
6。
癌症的预测不是患者水平。
-
7。
该方法不能预测特定的结果(即聚类或表型研究不符合资格)。
纵向预测指标被定义为允许不同时间点之间变化的变化的预测因子,即,仅在时间窗口内使用血液测试的最大值被视为纵向。并非所有预测因素都需要时间变化。如果作者描述了这些方法,则认为方法是AI或ML方法,或者这些方法比诸如逻辑回归之类的标准统计方法更为复杂。
数据提取和合成
提取了与文章的几个领域有关的数据:(1)出版物细节,例如作者,日记,日期;(2)研究特征,包括感兴趣的结果,研究设计,人口和设置以及样本量;(3)输入数据,包括所使用的数据类型和字段;(4)ML方法,包括任何功能工程,所使用的模型,评估指标和验证方法;(5)可重复性,包括数据和代码可用性;(6)使用Probast框架的质量评估[22]。Probast工具是评估预测模型中偏差风险的框架。该工具主要针对统计模型;但是,正在开发AI扩展名,但是,当前没有AI模型的等效工具[24]。因此,使用了原始的Probast工具。全文件2中提供了完整的提取表。
术语
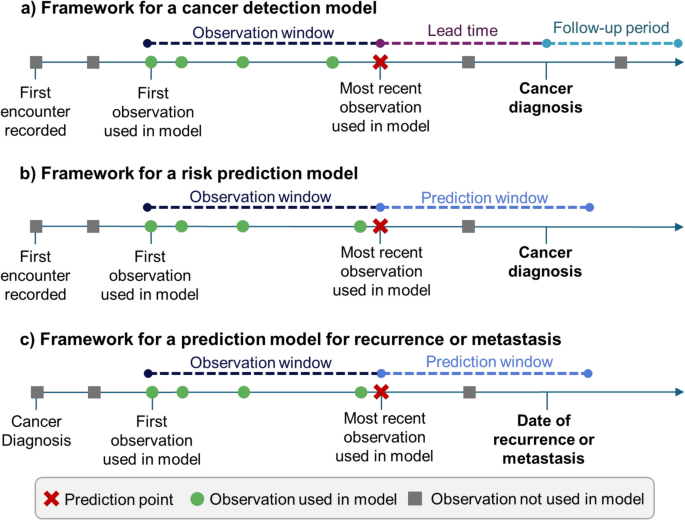
为了评估研究中研究中使用的模型,我们使用了[25]定义与每种模型相关的特定间隔:癌症检测模型,风险预测模型和转移/复发预测模型。该分类学在图2中证明。1。
癌症检测模型(图。1a)使用癌症患者的诊断日期,并定义对照组的索引日期。在观察窗口内,诊断或索引日期之前的数据用于预测结果。早期的癌症检测模型也应该有一个交货时间,这是模型的最后一个测量和结果之间的差距。在模型中未使用交货时间窗口中的数据。癌症检测模型可以包括减少偏见的随访期:这降低了对照癌症未发现的癌症的可能性。
风险预测模型(图1b)旨在寻找高风险人群。他们经常定义整个学习的时间框架,预测窗口,并在此窗口前使用数据来预测患者在此窗口内患癌症的风险。
预测复发或转移的模型更加多样化。他们可能会使用癌症最初诊断的监视数据来预测将来复发或转移的可能性(图。1c)。他们还可以在诊断之前使用数据。
[25],但是,对于此审查,需要少细胞分类。
结果
检索研究
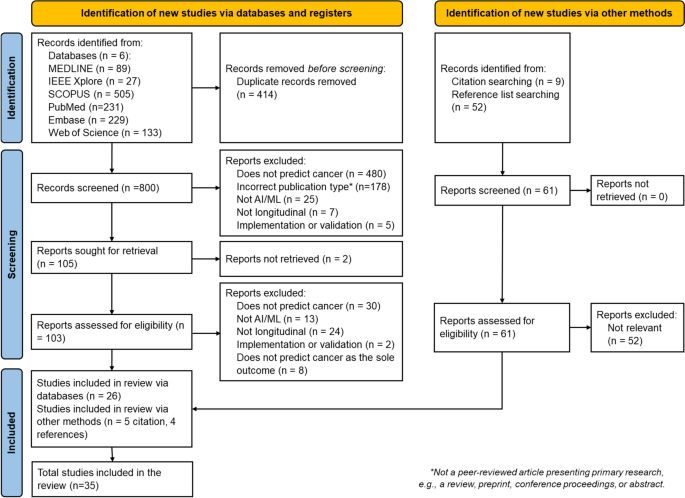
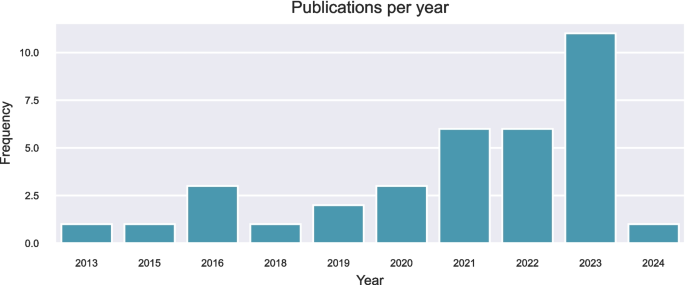
搜索6个数据库返回了1214项研究,其中414个是重复的。从参考和引文搜索中取回了另外61项研究。筛选和资格评估后,最终审查中包括35项研究。图2中提供了一个显示选择过程的流程图。2。按年发表的研究数量如图2所示。3。图2研究识别和选择的流程图。
21这是给出的图3
研究特征
研究环境和人口
研究使用了来自美国的人口(19,54%),荷兰(4,11%),台湾(5,14%),丹麦(2,6%),瑞典(1,3%),韩国(1,3%),以色列(1,3%)和新加坡(1,3%)。一项研究没有报告人口起源于何处,一项研究将来自英国的额外数据集作为验证集。五项研究(14%)使用单中心数据进行模型开发,20个(57%)使用了来自位置或医疗保健提供者连接的多个中心的数据,九项研究(25%)使用了全国范围的数据集,一项研究(3%)DID不报告研究设置。全国性的研究起源于瑞典,台湾,韩国和丹麦,而多中心的研究使用来自美国的附属实践的数据(n= 15),荷兰(n= 4)和以色列(n= 1)。研究使用了病例对照(9,26%),嵌套的病例对照(6,17%)或队列(20,57%)研究设计。在使用的数据集的设置中,四项研究使用了初级保健(11%),七个使用的二级护理(20%),23个使用了初级和二级护理数据(66%),一项没有报告。结果/预测任务
十项研究预测了特定时间范围内癌症的风险。
二十个研究重点是检测或早期检测癌症。一项研究预测转移,四项研究预测了复发。
研究中最常见的癌症是胰腺癌和结直肠癌(两项研究,26%)。有6项研究预测肺癌(17%),三项研究(9%)考虑肝癌和胃癌,分别考虑乳腺癌,皮肤,白血病和食管癌。一项研究分别预测了脑转移,小肠,肛门癌,宫颈癌和前列腺癌的癌症。此外,一项研究预测癌症是一种通用结果,没有指定地点。请注意,一些研究开发了多个站点的模型。
临床特征
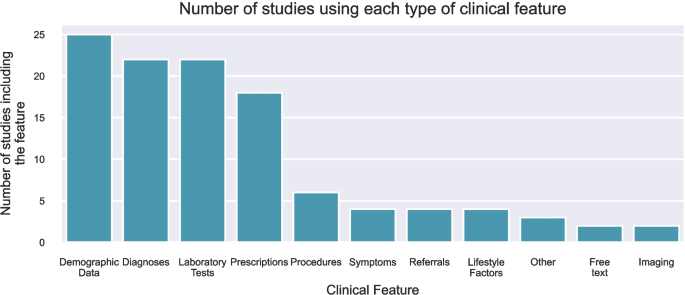
最常见的特征是人口统计学(25,71%),诊断(22,63%),实验室测试(22,63%)和处方(18,51%)。其他功能包括症状,推荐,程序,免费文本注释,生活方式因素,图像,肿瘤分期和组织学特征。特征的频率如图所示。4。
所选方法之间选择的临床变量有所不同。所有使用特征工程的研究都在其模型中使用了实验室测试,而仅使用顺序输入的模型中只有三分之一的模型也相同。此外,除了特征工程模型之一以外,所有使用人口统计数据都与三分之二(大约三分之二)相反(n= 12)顺序输入模型。模型特征
在预测模型中表示时间信息的方法可以分为两种主要方法。
第一种方法利用特征工程,即提取和操纵数据以形成信息变量的过程,以找到时间数据的有意义表示或捕获关键的时间特征。这些特征是为每个患者生成的,可以用作下游AI模型中的输入,以生成单个分类或风险评分。对于这种方法,我们分析了表示纵向信息的方法,而不是随后的AI模型,因为这些模型通常不是直接量身定制以处理纵向数据。第二种方法使用时间序列作为直接输入。这种方法通常包括一个具有特定机制来模拟顺序性质的模型,即时间步骤之间的依赖性。
用于表示顺序数据的功能工程
16个模型使用特征工程来表示时间数据。采取了各种方法;这些总结在表中1。
三项研究使用了趋势或斜率特征[26,,,,27,,,,28]。五种使用定义时间点之间的绝对变化,例如,预测日期之前的12个月更改[29,,,,30,,,,31,,,,32,,,,33]。对于其中两个研究[29,,,,30],从对每个患者训练该变量的单个轨迹的模型推断出用于计算绝对变化的值。Kinar等。[29]拟合线性回归模型,该模型用于预测指数前18和36个月的值,并且这些时间点之间的变化被用作趋势特征。Rodriguez等人也使用了类似的方法。[30],将线性混合效应模型拟合到对数转换的实验室测量中,以提供平滑的轨迹(即,测量误差减少)。然后使用预测来计算对数转换测量的6个月变化。
Rubenstein等。使用测量的最大增加和总变化为特征[34]。Beinecke等人使用了延迟动力学。al。[35]一项研究称使用了趋势特征,但没有描述如何计算这些特征[36]。在三项研究中,使用模式挖掘来找到预测的时间模式[37,,,,38,,,,39]。一项研究应用小波和傅立叶转换为纵向数据,并将系数用作模型的特征[40]。
无监督的方法还用于从时间序列中提取特征。Lasko等。和Ho等。两者都使用自动编码器学习患者轨迹的一般表示[40,,,,41]。
将顺序数据作为直接输入的模型
二十个研究使用了具有连续输入的深度学习模型,即原始序列或bined binned'分为离散的时间间隔。所使用的方法总结在表中2。
十项研究使用了基于复发性神经网络(RNN)的模型:七个使用了长期短期记忆模型(LSTMS)[40,,,,46,,,,47,,,,48,,,,49,,,,50,,,,51]和七个使用封闭的复发单位(GRU)[40,,,,46,,,,49,,,,50,,,,52,,,,53,,,,54,,,,55]。两项研究[53,,,,54]使用Choi等人提出的反向时间注意模型(保留)。这将注意力机制引入了GRU,以优先考虑患者的输入序列中最有意义的访问[56]。
五项研究使用了卷积神经网络(CNN)[40,,,,57,,,,58,,,,59,,,,60],一项研究[61]使用CNN-LSTM,代表诊断和药物为2D矩阵,并在输入上执行2D卷积。
三项研究[62,,,,63,,,,64]使用了标准的前馈神经网络,每个时间步长在体系结构中由一个节点表示。在其中两个,Park等人[63,,,,64]训练了每个变量的单独的神经网络,作为一个嵌入网络,以降低输入的维度,并将这些简化的功能串联以形成最终分类网络的输入。
六项研究利用了变压器体系结构[40,,,,49,,,,54,,,,55,,,,61,,,,65]。在一项研究中得出了位置编码,该研究使用了评估在输入序列中出现不同频率正弦函数的常见方法[40]。两项研究调整了这种方法,以便在患者的年龄中评估正弦功能,而不是在序列中的位置[49,,,,55]。Rasmy等。介绍多层嵌入到位置,不仅表示访问的顺序,还表示访问中的代码顺序[54]。两项研究没有报告嵌入的位置方法[61,,,,65]。
所使用的深度学习方法需要均匀长度的输入。有许多方法可以解决沿时间轴丢失的数据。五项研究具有代表特定窗口中事件的分类特征,因此输入的时间不需要特定注意[51,,,,58,,,,59,,,,60,,,,61]。如果使用事件表示为嵌入或数值值,则必须以某种方式强制最大序列长度短的任何序列。五项研究[40,,,,47,,,,48,,,,50,,,,65]``通过将零向量添加到序列中添加零向量来填充输入。三项研究[46,,,,52,,,,57]使用的前填充,通过使用最新目前的测量,沿时间轴丢失数据。六项研究没有报告解决输入序列长度的方法[54,,,,55,,,,62,,,,63,,,,64]。
预测窗口
表中显示了2.4中定义的每个模型的预测窗口3和4。桌子3显示了每个风险预测模型中使用的时间窗口。对于观察窗口,除一项风险预测研究外,所有其他人都使用了在研究期内的完整数据,并且在索引日期之前没有对数据施加任何限制。预测窗口在3到60个月之间变化,最常见的是36个月。只有一项风险预测研究调查了多个预测窗口[55]。这项研究介绍了他们关于36个月预测窗口的大多数结果,并指出这是一个合理的筛选窗口。
显示了20种癌症检测模型的时间窗口。其中五个使用患者的完整病史作为观察窗口。这些研究限制窗口的研究窗口在6到60个月之间。两项研究没有报告其观察窗口。一项研究并未按时间段定义他们的观察窗口,而是根据测量数量来定义他们的观察窗口。九项研究没有提前时间,而是使用癌症诊断前可用的所有数据来检测癌症。11项研究研究了使用交货时间早期检测到癌症的早期检测,这些研究在3到36个月之间有所不同。五项研究调查了许多交货时间。在大多数研究中,随访时间的报道很差,其中14项研究没有在此窗口上提供信息。四项研究没有进行诊断的随访对照,而两项研究给出了36个月的随访时间。
表中显示了转移和复发预测模型的窗口 5。一项研究包括对控制的随访[53]。两项研究将观察窗口的开始定义为与原发性癌症有关的特定临床事件[35,,,,40]。
与横截面模型进行比较
在本综述中包括的研究中,有7种比较了纵向方法与横截面方法,仅使用来自单个时间点的数据[26,,,,37,,,,38,,,,39,,,,52]。Ioannou等。报道使用顺序输入模型时,与横截面结果相比,歧视和校准的改善,但使用工程特征[没有显着改进[52]。Kop等。在一项设计的一项研究中,该研究具有改进的预测[37],但是在以后的工作中没有重复此结果[38]。Hoogendoorn等。没有报告预测性能的改善,但发现在敏感性分析的不同数据类型中,性能更稳定[39]。阅读等。注意到改进的趋势,但结果并不确定[26]。在Li等人的多模式研究中,发现纵向可以改善整合图像数据和临床数据的模态的预测,但并非所有模式[65]。
解释性
22个研究考虑了预测的解释性或模型推理。其中13个(10个特征工程模型,3个顺序输入模型)提出了模型级别的解释性,例如特征重要性,以证明该模型使用哪些信息来进行预测。九项研究(1个特征工程模型,8个顺序输入模型)具有预测水平的解释,其中计算出促成个人预测的因素。用于单个预测解释的方法是局部可解释的模型不合时宜的解释(lime)[47],基于注意力的解释[49,,,,53,,,,54],集成梯度[55,,,,61]和莎普利添加说明(Shap)[34,,,,36,,,,53,,,,64]。
研究的可重复性
13个研究(36%)有可在线使用的代码。两项研究使用了适合共同数据模型的数据:Kim等。使用了观察性医学结果伙伴关系共同数据模型(OMOP-CDM)[47]和Jia等。[28]使用的数据粘附到Trinetx标准数据模型[66]。一项研究[47]使用的数据可以在线免费获得。15项研究使用了可以要求或购买的数据集:退伍军人事务公司数据仓库[31,,,,32,,,,34,,,,52,,,,55],Kaiser Permanente南加州数据库[30,,,,31,,,,32,,,,33,,,,36],朱利叶斯一般从业者网络[38,,,,39,,,,46],Cerner健康事实[53,,,,54],HCUP状态住院数据库(SID)[50],IQVIA数据集[51]和trinetx [28]。六项研究使用的数据仅适用于原籍国的研究人员[27,,,,55,,,,58,,,,59,,,,61,,,,62]。
质量评估
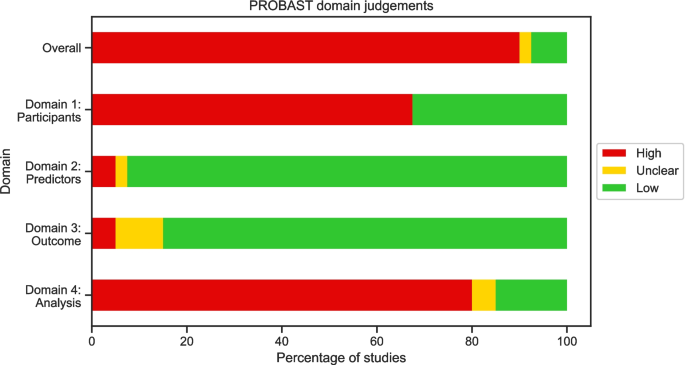
图2所示。5在附加文件2中提供了单个判断。在审查中,90%的研究的总体风险很高,低7.5%,而2.5%不清楚。
讨论
这项范围审查分析了适用于纵向EHR的AI方法,以预测癌症。确定了从纵向EHR中预测癌症的一系列方法。这些方法被分类为使用特征工程来表示时间数据以及直接使用顺序输入的方法。
该评论强调了使用该领域的常见方法和特征集,并且在研究之间的预测窗口中缺乏一致性。
评论的主要发现
全国范围内的研究是在瑞典,台湾,韩国和丹麦进行的,但是由于缺乏国家数据集,目前在许多国家不可能进行这些研究。所有使用来自多中心组织的数据的研究起源于三个国家:美国,荷兰和以色列。使用多中心数据和全国性数据,包括较大的数据集和结果的概括性,研究人员通常受到适当的数据集的限制,这是有好处的[8]。
将EHR数据用于预测模型的挑战已被充分记录[8],主要与数据质量问题和不同临床医生和网站之间的记录不一致有关[67]。Recording of data may also change over time within a healthcare centres, therefore additional care should be taken when developing longitudinal models to ensure models are robust to temporal shift [68]。While EHR poses extra challenges to analysis, if models are intended for use within EHR systems they are likely to encounter the same quality issues.Consideration of these aspects in model development should make the resulting algorithms more robust to similar issues upon deployment.EHR data provides numerous benefits over prospectively collected data as it is more reflective of clinical practice and is not as expensive or time-consuming to collect.
The intended use case of models is a key consideration when selecting data sources. If a model is intended to be used for early detection, this should be reflected in the dataset by utilising data that would be available at the point of use. Where studies are to use linked primary and secondary data, it should be considered whether these data would be linked in practice as this has implications for clinical applicability. However, proof of concept research demonstrating improved disease detection from linked data can still be valuable as it provides motivation for cohesive electronic health record systems across healthcare networks and many countries are aiming towards linked health data in practice.
The most frequently considered cancers were colorectal and pancreatic, accounting for more than 50% of the included studies. These are likely commonly chosen due to the impact they have globally; colorectal cancer is the third most common cancer and the second leading cause of cancer death. Pancreatic is less common, ranking around 12th, but contributes to the 6th largest number of deaths, and is known to be difficult to diagnose. There is an unmet need for earlier diagnosis of rarer cancers. Although more data is available for patients with more common cancers, there is an opportunity to establish methods on those datasets so they can then be implemented and optimised for rarer cancers.
The choice of features has an impact on the choice of model and vice versa—many of the approaches to feature engineering shown in Table 1, such as trend features and signal decomposition, would not be appropriate for categorical information such as diagnoses. Similarly, approaches to missing data differ between different types of variables; for categorical features, where the feature indicates whether the feature was present or not at that time, missing data do not need addressing, whereas for numerical features such as laboratory tests missing data must be imputed. This is particularly a problem in models requiring fixed inputs length inputs such as RNN based models and CNNs.
The methods identified in this review are summarised along with their advantages and limitations in Tables1和2分别。The most commonly used feature engineering method was absolute change in measurement, which is likely commonly chosen due to the ease of computation but requires expert knowledge to determine which times to calculate change between.The most common approach using sequential inputs was to use models based on RNNs.A general advantage of feature engineering is that the features can be used in relatively simple artificial intelligence algorithms, reducing the computational cost, although they require human input in crafting meaningful features.Alternatively, deep learning approaches have the capability to learn hidden patterns without the need for explicit crafting by an expert, including potentially undiscovered predictors.This gives rise to a key question;does added complexity increase accuracy, and does this increase justify the increase cost.The two approaches are rarely compared, and future research should aim to do this.
In addition, research should consider whether longitudinal data does improve the predictive capability of models. Few studies in this review compared longitudinal models to cross-sectional approaches, and those that did were not definitive in finding an improvement in performance although there was weak evidence to support an improvement, and no studies reported that longitudinal data harms predictions. Given the additional complexity and cost of incorporating longitudinal data, the question of whether this is justified should be considered.
As previously described, longitudinal data in healthcare provide specific challenges for prediction models. The methods found in this review address these in varying ways. Data irregularity was commonly addressed in feature engineering models by modelling patients’ trajectories individually to infer values at specific time-points or by calculating slopes from available data. Sequential methods often coded the relative times of observations to provide context to the models or required direct imputation of missing data in the temporal axis. Data opacity was considered in a number of studies aiming to develop explainable methods. The level of explainability achieved by models varied by the approach taken; feature engineering models were more likely to provide model level explanations, which are often simple to implement. However, these may not be as useful for clinicians as prediction level explanations, which can help a user understand why the model classified a patient a certain way, but do require more complex methods to implement, increasing the computational cost of a model.
Risk prediction models were evaluated against the longitudinal model framework described in Fig. 1。All risk prediction models except one used the full study period as the observation window, and no studies evaluated models using different observation windows.In risk predictions studies, this was generally a universal time window for the entire cohort, for example, from 2003–2011.Using all available data as the observation window may result in better performance as the full history is used, providing more context for patient data.Conversely, using all available data may hinder performance, by introducing additional noise into models.In addition, using longer time sequences may increase complexity of models and increase computational expense.Given this potential trade-off, studies should aim to evaluate the impact of various observation windows on model performance.Similarly, only one study experimented with different prediction windows.The risk prediction windows used by other studies varied significantly, even when predicting the same cancer, suggesting there is not clear window that should be assumed without investigation.
Cancer detection models should report three quantities: the observation window, lead time window, and follow-up time. A number of studies used the full patient history as the observation window, which has the potential to introduce bias to models as cancer patients may have systematically shorter trajectories as a result, which may be detected by sequential input models. Potential bias should also be mitigated by ensuring there is sufficient follow up of the control population, as patients may have been diagnosed with the cancer of interest at a later date, indicating a present but as yet undiagnosed cancer. Only one study reported including any follow up time [27]。Lead time is a key parameter to consider in early detection models.Most early detection studies experimented with different lead times, which allows for interpretation of how prediction accuracy changes with distance from the event of interest.
In general, the reporting of time-windows was poor in metastasis and recurrence prediction models. This makes it difficult to not only assess potential bias in the models, but also makes the intended use-case unclear, i.e., where would the prediction be made and how would this aid a clinician. As previously explained, follow-up time should be reported in studies predicting recurrence or metastasis to rule out potentially undiagnosed patients and hence mislabelled occurrences.
Given that current research into the use of longitudinal health records is in the early stages and studies are generally proof-of-concept, the reproducibility of the research is vital to ensure future work can build upon findings. Despite this, only around a third of studies included in the review have code that is available. Due to the confidential nature of health data, open access data is rare, however the availability of commercial datasets such as those used by studies in this review provides the opportunity for comparative works. For research using these sources it is especially important to be clear about how cohorts were selected. Clear reporting of methods and study setting is vital for reproducibility. The recent publication of an AI extension to the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD-AI) statement provides a checklist of reporting items that should be followed by predictive models using AI [69]。
The PROBAST assessment showed that most studies were at high risk of bias, with only three studies achieving low risk of bias overall. This is unsurprising given that the studies are generally exploratory, however the results highlight the common areas where risk of bias is introduced. The highest risk domains were domains 1 and 4, concerning study participants and analysis respectively. High risk in domain 1 was introduced due to case–control study designs and restrictions on participants (e.g., age based) without acknowledgement of how this affects the applicability of the model. These factors may be unavoidable due to data access restrictions; however, researchers should make the potential impact on the risk of bias clear and nested case–control studies should adjust for outcome frequency as described in the PROBAST framework [22]。In domain 4, common areas introducing risk of bias were an insufficient number of participants with the outcome, lack of follow-up periods for controls, inappropriate performance measures, and no accounting for overfitting.While low numbers of patients with the outcome is often determined by available data, the remaining areas can be mitigated by researchers through the following actions: ensuring follow-up of controls;reporting comprehensive performance measures, including both discrimination and calibration measures;and using cross-validation or bootstrapping to account for overfitting in the model.
未来工作的建议
In conducting this review, we identified several common areas for improvement which future work should aim to address:
-
1。
Models should be reported using clear terminology, provided here or by Lauritsen et al.[25] Given the clinical application, it is especially important to clearly explain at which point the prediction model would be used, and which data would be available.
-
2。
Given the lack of consensus on appropriate prediction windows for each of the cancers, studies should evaluate models at various time points to assess the optimal time windows for prediction. In addition, models should be evaluated against cross-sectional methods. It is not a given that adding longitudinal data improves performance, but it is likely to increase complexity. Researchers should aim to evaluate whether this added expense is justified for the problem.
-
3。
To ensure reproducibility of research, studies using AI for prediction of cancer should adhere to the TRIPOD + AI statement to ensure methods are transparent [69]。Where possible, data should be made available for sharing.As this is often not appropriate for patient data, reporting of datasets should be comprehensive.Researchers should make the code used in the research publicly available.
-
4。
When conducting cancer prediction research, researchers should be mindful of how bias may be introduced to the model. The forthcoming PROBAST-AI will provide guidance [24]。Mitigation strategies include ensuring sufficient follow-up of controls;reporting a variety of performance measures, including discrimination and calibration;and accounting for optimism and overfitting in the model using cross-validation or bootstrapping.
Strengths of the current study
This review has multiple strengths. Firstly, the scope of the review covered all longitudinal methods to include a wide range of methodologies, found through an exhaustive search strategy. In addition, the review adheres to the PRISMA guidance for conducting scoping reviews. The review also provides a PROBAST assessment, highlighting common areas where risk of bias is high in longitudinal models.
Limitations of the current study
This review has four main limitations:
-
1。
Firstly, the records were only screened by one author. The impact of this was mitigated by taking a lenient approach to inclusion; articles were only excluded initially if the author was confident in their ineligibility. Where this was not the case, fellow authors were consulted to reach a consensus.
-
2。
Secondly, a number of studies included in this review were found via citation and reference searching and were not captured as part of the initial search strategy. These studies were missed by the search strategy due to several factors; some did not state in the title or abstract that they included temporal data while some did not mention health records. Two terms were identified from these results that can be used to describe longitudinal data: ‘sequential’ and ‘trajectory’. While we are confident the most significant studies in the area were found, inclusion of these terms could have made the search strategy more comprehensive.
-
3。
This review did not quantify the retrieved works as precisely as the framework described by Lauritsen [25] as this granularity would have impeded the ability to compare similar studies.
-
4。
Finally, this review does not comment on the relative performances of each of the methods due to the heterogeneity of applications and datasets. The review can also not provide an answer as to whether longitudinal methods improve upon cross-sectional methods as this was rarely evaluated in the studies and is likely to be problem dependent.
结论
这篇评论发现,已应用于纵向EHR的一系列技术,包括趋势特征的工程和基于RNN的模型。这些模型用于预测一系列癌症,但最常见的是胰腺癌或结直肠癌。最大程度地降低偏见的风险对于确保在可部署研究方面的进步至关重要。发现潜在偏见的关键领域,通常与选择队列和数据分析有关。这些潜在的缓解策略包括对控制人群的足够随访以及对模型绩效的强大评估。该领域的未来研究应旨在评估使用一系列时间窗口的预测模型,以找到用于应用模型的最佳时间表;该分析在检索的研究中很少存在。为了帮助这些模型从探索性研究到临床实践的发展,研究人员应旨在清楚地报告方法,遵守可用的分类法和报告指南。
数据可用性
The dataset supporting the conclusions of this article are included within the article and its additional files.
参考
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al.2022年全球癌症统计:Globocan在185个国家 /地区的36个癌症全球发病率和死亡率的估计。CA: A Cancer Journal for Clinicians.2024;74(3):229–63.eprint: https://onlinelibrary.wiley.com/doi/pdf/https://doi.org/10.3322/caac.21834。World Health Organisation.Guide to cancer early diagnosis.Geneva: World Health Organisation; 2017.Available from:
https://www.who.int/publications-detail-redirect/9789241511940。Hamilton W, Walter FM, Rubin G, Neal RD.Improving early diagnosis of symptomatic cancer.
Nature Reviews Clinical Oncology.2016 Dec;13(12):740–9.Publisher: Nature Publishing Group.可从:https://www.nature.com/articles/nrclinonc.2016.109。Vickers AJ, Brewster SF.
PSA Velocity and Doubling Time in Diagnosis and Prognosis of Prostate Cancer.Brit J Med Surg Urol.2012;5(4):162–8.Publisher: SAGE Publications.可从:https://doi.org/10.1016/j.bjmsu.2011.08.006。Hunter B, Hindocha S, Lee RW.
The Role of Artificial Intelligence in Early Cancer Diagnosis.癌症。2022 Mar;14(6):1524.可从:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8946688/。IBM。
What is Artificial Intelligence (AI)?;2023. Available from:https://www.ibm.com/topics/artificial-intelligence。International Organization for Standardisation, International Electrotechnical Commision. ISO/IEC 22989:2022(en), Information technology — Artificial intelligence — Artificial intelligence concepts and terminology; 2022. Available from:
https://www.iso.org/obp/ui/en/#iso:std:iso-iec:22989:ed-1:v1:en:term:3.1.4。Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA.Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review.
Journal of the American Medical Informatics Association : JAMIA.2017 Jan;24(1):198.Publisher: Oxford University Press.可从:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5201180/。Pereira SP, Oldfield L, Ney A, Hart PA, Keane MG, Pandol SJ, et al.
Early detection of pancreatic cancer.The Lancet Gastroenterology & Hepatology.2020;5(7):698–710.Publisher: Elsevier.可从:https://www.thelancet.com/journals/langas/article/PIIS2468-1253(19)30416-9/fulltext。Pannala R, Basu A, Petersen GM, Chari ST.
New-onset Diabetes: A Potential Clue to the Early Diagnosis of Pancreatic Cancer.The lancet oncology.2009 Jan;10(1):88–95.可从:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2795483/。Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer. A meta-analysis JAMA. 1995;273(20):1605–9.
CAS
一个 PubMed一个 Google Scholar一个 Co¸skun A, Carobene A, Kilercik M, Serteser M, Sandberg S, Aarsand AK, et al.Within-subject and between-subject biological variation estimates of 21 hematological parameters in 30 healthy subjects.
Clinical Chemistry and Laboratory Medicine (CCLM).2018 Aug;56(8):1309–18.Publisher: De Gruyter.可从:https://doi.org/10.1515/cclm-2017-1155/html?lang=en。Che Z, Purushotham S, Cho K, Sontag D, Liu Y. Recurrent Neural Networks for Multivariate Time Series with Missing Values.
科学报告。2018 Apr;8(1):6085.Publisher: Nature Publishing Group.可从:https://www.nature.com/articles/s41598-018-24271-9。Zhang D, Yin C, Hunold KM, Jiang X, Caterino JM, Zhang P. An interpretable deep-learning model for early prediction of sepsis in the emergency department.
模式。2021;2(2).可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-85100825661&doi=10.1016%2fj.patter.2020.100196&partnerID=40&md5=ddd497778d69d1f5f94fcd2e1d67ddba。Zargoush M, Sameh A, Javadi M, Shabani S, Ghazalbash S, Perri D. The impact of recency and adequacy of historical information on sepsis predictions using machine learning.
科学报告。2021 Oct;11(1):20869.Publisher: Nature Publishing Group.可从:https://www.nature.com/articles/s41598-021-00220-x。Ismail Fawaz H, Forestier G, Weber J, Idoumghar L, Muller PA.
Deep learning for time series classification: a review.Data Mining and Knowledge Discovery.2019 Jul;33(4):917–63.可从:https://doi.org/10.1007/s10618-019-00619-1。Wang Q, Farahat A, Gupta C, Zheng S. Deep time series models for scarce data.
Neurocomputing.2021 Oct;456:504–18.可从:https://www.sciencedirect.com/science/article/pii/S0925231221001922。Xie F, Yuan H, Ning Y, Ong MEH, Feng M, Hsu W, et al.
Deep learning for temporal data representation in electronic health records: A systematic review of challenges and methodologies.Journal of Biomedical Informatics.2022;126.可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-85122621542&doi=10.1016%2fj.jbi.2021.103980&partnerID=40&md5=65386bfe354ffab69e1846c9fd026a61。Carrasco-Ribelles LA, Llanes-Jurado J, Gallego-Moll C, Cabrera-Bean M, Monteagudo-Zaragoza M, Viol´an C, et al.
Prediction models using artificial intelligence and longitudinal data from electronic health records: a systematic methodological review.Journal of the American Medical Informatics Association.2023 Dec;30(12):2072–82.可从:https://doi.org/10.1093/jamia/ocad168。Cascarano A, Mur-Petit J, Hern´andez-Gonz´alez J, Camacho M, de Toro Eadie N, Gkontra P, et al. Machine and deep learning for longitudinal biomedical data: a review of methods and applications. Artificial Intelligence Review. 2023 Aug. Available from:
https://doi.org/10.1007/s10462-023-10561-w。Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al.Prisma 2020声明:报告系统评价的最新指南。
BMJ (Clinical research ed).2021;372: n71.
PubMed一个 Google Scholar一个
Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: A Tool to Assess Risk of Bias and Applicability of Prediction Model Studies: Explanation and Elaboration. Annals of Internal Medicine. 2019 Jan;170(1):W1-W33. Publisher: American College of Physicians. Available from: https://www.acpjournals.org/doi/abs/https://doi.org/10.7326/M18-1377。Andrea C Tricco, Erin Lillie, Wasifa Zarin, Kelly K O’Brien, Heather Colquhoun, Danielle Levac, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Annals of Internal Medicine. 2018;169(7):467–73. Available from: https://www.acpjournals.org/doi/
https://doi.org/10.7326/M18-0850。Collins GS, Dhiman P, Navarro CLA, Ma J, Hooft L, Reitsma JB, et al.Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence.
BMJ开放。2021 Jul;11(7):e048008.Publisher: British Medical Journal Publishing Group Section: Medical publishing and peer review.可从:https://bmjopen.bmj.com/content/11/7/e048008。Lauritsen SM, Thiesson B, Jørgensen MJ, Riis AH, Espelund US, Weile JB, et al.
The Framing of machine learning risk prediction models illustrated by evaluation of sepsis in general wards.NPJ数字医学。2021 Nov;4(1):1–12.Publisher: Nature Publishing Group.可从:https://www.nature.com/articles/s41746-021-00529-x。Read AJ, Zhou W, Saini SD, Zhu J, Waljee AK.
Prediction of Gastrointestinal Tract Cancers Using Longitudinal Electronic Health Record Data.癌症。2023;15(5).
Cichosz SL, Jensen MH, Hejlesen O, Henriksen SD, Drewes AM, Olesen SS.Prediction of pancreatic cancer risk in patients with new-onset diabetes using a machine learning approach based on routine biochemical parameters.Computer Methods and Programs in Biomedicine.2024 Feb;244:107965.可从:https://www.sciencedirect.com/science/article/pii/S0169260723006314。Jia K, Kundrot S, Palchuk MB, Warnick J, Haapala K, Kaplan ID, et al.
A pancreatic cancer risk prediction model (Prism) developed and validated on large-scale US clinical data.eBioMedicine.2023 Dec;98.Publisher: Elsevier.可从:https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(23)00454-1/fulltext。Kinar Y, Kalkstein N, Akiva P, Levin B, Half EE, Goldshtein I, et al.
Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: a binational retrospective study.Journal of the American Medical Informatics Association.2016 Sep;23(5):879–90.可从:https://academic.oup.com/jamia/article/23/5/879/2379871。Rodriguez PJ, Heagerty PJ, Clark S, Khor S, Chen Y, Haupt E, et al. Using Machine Learning to Leverage Biomarker Change and Predict Colorectal Cancer Recurrence. JCO Clinical Cancer Informatics. 2023;7: e2300066.
文章
一个 PubMed一个 PubMed Central一个 Google Scholar一个 Chen W, Zhou Y, Xie F, Butler RK, Jeon CY, Luong TQ, et al.Derivation and External Validation of Machine Learning-Based Model for Detection of Pancreatic Cancer.
Am J Gastroenterol。2023;118(1):157–67.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Chen W, Zhou B, Jeon CY, Xie F, Lin YC, Butler RK, et al. Machine learning versus regression for prediction of sporadic pancreatic cancer. Pancreatology. 2023;23:396–402.
文章一个 PubMed一个 Google Scholar一个
Chen W, Butler RK, Lustigova E, Chari ST, Maitra A, Rinaudo JA, et al. Risk Prediction of Pancreatic Cancer in Patients With Recent-onset Hyperglycemia: A Machine-learning Approach. J Clin Gastroenterol. 2023;57(1):103–10.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Rubenstein JH, Fontaine S, MacDonald PW, Burns JA, Evans RR, Arasim ME, et al.Predicting Incident Adenocarcinoma of the Esophagus or Gastric Cardia Using Machine Learning of Electronic Health Records.胃肠病学。2023;165(6):1420-9.e10.
文章一个 PubMed一个 Google Scholar一个
Beinecke JM, Anders P, Schurrat T, Heider D, Luster M, Librizzi D, et al.Evaluation of machine learning strategies for imaging confirmed prostate cancer recurrence prediction on electronic health records.Comput Biol Med。2022;143: 105263.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Gould MK, Huang BZ, Tammemagi MC, Kinar Y, Shiff R. Machine Learning for Early Lung Cancer Identification Using Routine Clinical and Laboratory Data.American Journal of Respiratory and Critical Care Medicine.2021 Aug;204(4):445–53.可从:https://doi.org/10.1164/rccm.202007-2791OC。Kop R, Hoogendoorn M, Moons LMG, Numans ME, ten Teije A. On the advantage of using dedicated data mining techniques to predict colorectal cancer.
In: Holmes JH, Bellazzi R, Sacchi L, Peek N, editors.Artificial Intelligence in Medicine.卷。9105 of Lectures in Computer Science.Cham: Springer International Publishing;2015. p.13342. Available from:https://www.scopus.com/inward/record.uri?eid=2-s2.0-84947998729&doi=10.1007%2f978-3-319-19551-316&partnerID=40&md5=0a8da5ef9f24a88fd30fed6d97ae7e51。Kop R, Hoogendoorn M, Teije AT, Bu¨chner FL, Slottje P, Moons LMG, et al.
Predictive modeling of colorectal cancer using a dedicated pre-processing pipeline on routine electronic medical records.Computers in Biology and Medicine.2016;76:30–8.可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-84977074295&doi=10.1016%2fj.compbiomed.2016.06.019&partnerID=40&md5=94d1281b295d78adf6037c8f28a970a3。Hoogendoorn M, Szolovits P, Moons LMG, Numans ME.
Utilizing uncoded consultation notes from electronic medical records for predictive modeling of colorectal cancer.Artif Intell Med。2016;69:53–61.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ho D, Tan IBH, Motani M. Predictive models for colorectal cancer recurrence using multi-modal healthcare data. In: Proceedings of the Conference on Health, Inference, and Learning. Virtual Event USA: ACM; 2021. p. 204–13. Available from: https://dl.acm.org/doi/https://doi.org/10.1145/3450439.3451868。Lasko T, Denny J, Levy M. Computational Phenotype Discovery Using Unsupervised Feature Learning over Noisy, Sparse, and Irregular Clinical Data.
PLOS一个。2013;8(6).
Bishop CM.Neural Networks.In: Pattern Recognition and Machine Learning.New York, NY: Springer;2006. p.225–90.可从:https://doi.org/10.1007/978-0-387-45528-05。Bishop CM, Bishop H. Transformers.
In: Bishop CM, Bishop H, editors.Deep Learning: Foundations and Concepts.Cham: Springer International Publishing;2024. p.357–406.可从:https://doi.org/10.1007/978-3-031-45468-412。Zhao B, Lu H, Chen S, Liu J, Wu D. Convolutional neural networks for time series classification.
Journal of Systems Engineering and Electronics.2017;28(1):162–9.可从:https://ieeexplore.ieee.org/document/7870510。Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al.
Attention is All you Need.In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17).卷。30. Long Beach, CA, USA: Curran Associates, Inc.;2017. Available from:https://proceedings.neurips.cc/paperfiles/paper/2017/hash/3f5ee243547dee91fbd053c1c4a845aa-Abstract.html。Amirkhan R, Hoogendoorn M, Numans ME, Moons L. Using recurrent neural networks to predict colorectal cancer among patients.
In: 2017 IEEE Symposium Series on Computational Intelligence (SSCI).卷。2018-January;2018. p.1–8.可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-85046102195&doi=10.1109%2fSSCI.2017.8280826&partnerID=40&md5=b51d6347d8f131ba7773e399dbd6adcc。Kim HH, Lim YS, Seo SI, Lee KJ, Kim JY, Shin WG.
A Deep Recurrent Neural Network-Based Explainable Prediction Model for Progression from Atrophic Gastritis to Gastric Cancer.应用科学。2021 Jan;11(13):6194.Number: 13 Publisher: Multidisciplinary Digital Publishing Institute.可从:https://www.mdpi.com/2076-3417/11/13/6194。Sanyal J, Tariq A, Kurian AW, Rubin D, Banerjee I. Weakly supervised temporal model for prediction of breast cancer distant recurrence.
科学报告。2021;11(1).可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-85105250713&doi=10.1038%2fs41598-021-89033-6&partnerID=40&md5= 241297d4a9dc64bfec263bd8b5cb7b45.
Zhang Y, Hu C, Zhong L, Song Y, Sun J, Li M, et al. Spatiotemporal Attention for Early Prediction of Hepatocellular Carcinoma Based on Longitudinal Ultrasound Images. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. Lecture Notes in Computer Science. Cham: Springer Nature Switzerland; 2022. p. 534–43.
Andjelkovic J, Ljubic B, Ameen Abdel Hai, Stanojevic M, Diaz W, Obradovic Z. Sequential machine learning in prediction of common cancers.医学信息学解锁。2022;30.可从:https://www.sciencedirect.com/science/article/pii/S2352914822000764。Wang Y, Wu T, Wang Y, Wang G. Enhancing Model Interpretability and Accuracy for Disease Progression Prediction via Phenotype-Based Patient Similarity Learning.
In: Biocomputing 2020. World Scientific;2019. p.511–22.可从:https://doi.org/10.1142/97898112156360045。Ioannou GN, Tang W, Beste LA, Tincopa MA, Su GL, Van T, et al.
Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients With Hepatitis C Cirrhosis.JAMA network open.2020;3(9):e2015626.可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0-85090180783&doi=10.1001%2fjamanetworkopen.2020.15626&partnerID=40&md5=1ce254174206164f1e4cacd484f84f6c。Li Z, Li R, Zhou Y, Rasmy L, Zhi D, Zhu P, et al. Prediction of Brain Metastases Development in Patients With Lung Cancer by Explainable Artificial Intelligence From Electronic Health Records. JCO clinical cancer informatics. 2023 Apr;7:e2200141. Place: United States.
L Rasmy Y Xiang Z Xie C Tao D Zhi Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction.
NPJ数字医学。2021;4(1):86 Available from:https://doi.org/10.1038/s41746-021-00455-y。Placido D, Yuan B, Hjaltelin JX, Zheng C, Haue AD, Chmura PJ, et al.
A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories.自然医学。2023 May;29(5):1113–22.Number: 5 Publisher: Nature Publishing Group.可从:https://www.nature.com/articles/s41591-023-02332-5。Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. RETAIN: An Interpretable Predictive Model for Healthcare using Reverse Time Attention Mechanism.
In: Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS 2016).巴塞罗那;2016. p.3512–20.
Gillstedt M, Polesie S. Ability to Predict Melanoma Within 5 Years Using Registry Data and a Convolutional Neural Network: A Proof of Concept Study. Acta Dermato-Venereologica. 2022 Jul;102:adv00750.
Wang YH, Nguyen PA, Islam MM, Li YC, Yang HC. Development of Deep Learning Algorithm for Detection of Colorectal Cancer in EHR Data. Studies in Health Technology and Informatics. 2019;264:438–41.
PubMed一个 Google Scholar一个
Yeh MCH, Wang YH, Yang HC, Bai KJ, Wang HH, Li YCJ.Artificial Intelligence–Based Prediction of Lung Cancer Risk Using Nonimaging Electronic Medical Records: Deep Learning Approach.Journal of Medical Internet Research.2021 Aug;23(8):e26256.可从:https://www.jmir.org/2021/8/e26256。Wang HH, Wang YH, Liang CW, Li YC.Assessment of Deep Learning Using Nonimaging Information and Sequential Medical Records to Develop a Prediction Model for Nonmelanoma Skin Cancer.
JAMA Dermatology.2019 Nov;155(11):1277–83.可从:https://doi.org/10.1001/jamadermatol.2019.2335。Chen HY, Wang HM, Lin CH, Yang R, Lee CC.
Lung Cancer Prediction Using Electronic Claims Records: A Transformer-Based Approach.IEEE Journal of Biomedical and Health Informatics.2023 Dec;27(12):6062–73.Conference Name: IEEE Journal of Biomedical and Health Informatics.可从:https://ieeexplore.ieee.org/document/10283843。Hung CY, Chen HY, Wee LJK, Lin CH, Lee CC.
Deriving A Novel Health Index Using A Large-Scale Population Based Electronic Health Record With Deep Networks.In: IEEE Engineering in Medicine and Biology Society Conference Proceedings.IEEE Engineering in Medicine and Biology Society Conference Proceedings.IEEE;2020年。5872–5.Place: 345 E 47TH ST, NEW YORK, NY 10017 USA.
Park J, Artin MG, Lee KE, Pumpalova YS, Ingram MA, May BL, et al.Deep learning on time series laboratory test results from electronic health records for early detection of pancreatic cancer.Journal of Biomedical Informatics.2022;131.可从:https://www.scopus.com/inward/record.uri?eid=2-s2.0–85131095978&doi=10.1016%2fj.jbi.2022.104095&partnerID=40&md5=558d243ffd56c91d2cdcb85e191d198c。Park J, Artin MG, Lee KE, May BL, Park M, Hur C, et al. Structured deep embedding model to generate composite clinical indices from electronic health records for early detection of pancreatic cancer. Patterns (New York, NY). 2023;4(1): 100636.
CAS
一个 Google Scholar一个 Li TZ, Still JM, Xu K, Lee HH, Cai LY, Krishnan AR, et al. Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures from Routine EHRs for Pulmonary Nodule Classification. In: Greenspan H, Madabhushi A, Mousavi P, Salcudean S, Duncan J, Syeda-Mahmood T, et al., editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. Cham: Springer Nature Switzerland; 2023. p. 649–59.章一个
Google Scholar
一个 Palchuk MB, London JW, Perez-Rey D, Drebert ZJ, Winer-Jones JP, Thompson CN, et al.A global federated real-world data and analytics platform for research.JAMIA Open.
2023 May;6(2):ooad035.可从:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10182857/。Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. Journal of the American Medical Informatics Association: JAMIA. 2013;20(1):144–51.
文章
一个 PubMed一个 PubMed Central一个 Google Scholar一个 Guo LL, Pfohl SR, Fries J, Posada J, Fleming SL, Aftandilian C, et al.Systematic Review of Approaches to Preserve Machine Learning Performance in the Presence of Temporal Dataset Shift in Clinical Medicine.
Applied Clinical Informatics.2021 Sep;12:808–15.Publisher: Georg Thieme Verlag KG.可从:https://doi.org/10.1055/s-0041-1735184。Collins GS, Moons KGM, Dhiman P, Riley RD, Beam AL, Calster BV, et al.
TRIPOD+AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods.BMJ。2024 Apr;385:e078378.Publisher: British Medical Journal Publishing Group Section: Research Methods & Reporting.可从:https://www.bmj.com/content/385/bmj-2023-078378。下载参考
Code used for searching each database is provided in Additional File 1.
资金
This research was financially supported by the UK Research and Innovation Engineering and Physical Sciences Research Council (grant number EP/S024336/1/).
作者信息
不适用。
同意出版
不适用。
竞争利益
作者没有宣称没有竞争利益。
附加信息
出版商的注释
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。
补充信息
12874_2025_2473_MOESM5_ESM.xlsx
Additional File 5. This file contains the extracted data items describing the feature engineering and sequential input methods of the studies.
权利和权限
开放访问
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
本文中的图像或其他第三方材料包含在文章的Creative Commons许可中,除非在材料的信用额度中另有说明。如果文章的创意共享许可中未包含材料,并且您的预期用途不得由法定法规允许或超过允许的用途,则需要直接从版权所有者那里获得许可。要查看此许可证的副本,请访问http://creativecommons.org/licenses/4.0/。重印和权限
引用本文
Moglia, V., Johnson, O., Cook, G.
等。Artificial intelligence methods applied to longitudinal data from electronic health records for prediction of cancer: a scoping review.BMC Med Res Methodol25 , 24 (2025). https://doi.org/10.1186/s12874-025-02473-w下载引用
:2024年5月30日
:2025年1月17日
:2025年1月28日
:https://doi.org/10.1186/s12874-025-02473-w关键字