为生物医学自然语言处理应用程序和建议基准测试大型语言模型
作者:Xu, Hua
介绍
生物医学文献由于其巨大的体积和特定于领域的挑战而引起了策展,解释和知识发现的直接障碍。仅PubMed就会看到每天增加约5000篇文章,截至2024年3月,总计超过3600万。1。在Covid-19等专业领域,每月添加大约10,000篇专用文章,截至2024年3月,总数超过40万2。除了数量外,生物医学领域还用模棱两可的语言提出了挑战。例如,可以使用763个不同的术语引用一个单一的实体,例如长covid3。此外,同一术语可以描述不同的实体,如术语AP2所示,该实体可以指基因,化学或细胞系4。除了实体之外,识别新颖的生物医学关系并捕获生物医学文献中的语义提出了进一步的挑战5,,,,6。为了克服这些挑战,使用生物医学自然语言处理(BIONLP)技术来协助手动策划,解释和知识发现。生物医学模型被认为是Bionlp方法的骨干。
他们利用大量的生物医学文献,并以无监督或自制的方式捕获生物医学语义表示。早期生物医学语言模型是使用完全连接的神经网络(例如biowordvec和biosentvec)的非上下文嵌入(例如Word2Vec和fastText)4,,,,7,,,,8。自从变形金刚诞生以来,生物医学模型已经采用了它们的体系结构,并且可以使用来自变形金刚体系结构的编码器(例如生物医学双向编码器)(包括Biobert和PubMedbert和PubMedbert)等变形金学架构的编码器分类为(1)基于编码器的掩盖语言模型9,,,,10,,,,11,(2)基于解码器的生成语言模型,使用来自变压器体系结构的解码器,例如生成培训的变压器(GPT)家族,包括MioGPT和BioMedLM12,,,,13,以及(3)基于编码器,使用编码器和解码器,例如Biobart和Scifive14,,,,15。Bionlp研究微调了这些语言模型,并证明它们在各种Bionlp应用中实现了SOTA性能10,,,,16,这些模型已成功地用于PubMed尺度下游应用程序,例如生物医学句子搜索17和COVID-19文献挖掘2。
最近,包括GPT-3和更值得注意的GPT-4在内的最新封闭源GPT模型已取得了很大的进步,并引起了社会的大量关注。这些模型的关键特征是其参数的指数增长。例如,GPT-3具有约1750亿个参数,比GPT-2大。这种大小的模型通常称为大语言模型(LLM)18。此外,LLM的增强是通过通过人类的反馈来加强学习来实现的,从而使文本生成与人类的偏好保持一致19。例如,GPT-3.5使用强化学习技术建立在GPT-3的基础上,从而显着提高了自然语言理解的性能20。使用GPT-3.5和GPT-4的Chatgpt启动聊天机器人标志着生成人工智能的里程碑。它在其前任未能执行的任务中证明了强大的能力。例如,GPT-4通过了20多次学术和专业考试,包括统一的律师考试,SAT基于证据的阅读和写作以及医学知识自我评估计划21。出色的进步引发了社会之间的广泛讨论,引起了人们的兴奋和关注。除了封闭源LLM,开源LLM,例如Llama22和混合23在下游应用程序中已被广泛采用,也用作连续预处理的特定领域资源的基础。在生物医学领域中,PMC Llama(7b和13b)是第一个生物医学领域特异性的LLMS之一,在4.8的生物医学论文和30 K医学教科书上不断预先培训的Llama24。Meditron(7b和70b)是最新的生物医学特异性LLM,在Llama 2上采用了类似的连续预处理策略。
开拓性研究已经对生物医学领域的LLM进行了早期实验,并报告了令人鼓舞的结果。例如,Bubeck等。研究了GPT-4在广泛的范围中的能力,例如编码,数学和与人的互动。这项早期研究报告了与生物医学相关的结果,表明GPT-4在美国医学许可检查中获得了约80%的准确性(步骤1、2和3),以及使用GPT-4在医疗票据中验证索赔的示例。Lee等。还展示了GPT-4的用例,用于回答医疗问题,从患者报告中产生摘要,协助临床决策并创建教育材料24。Wong等。对GPT-3.5和GPT-4进行了针对端到端临床试验匹配,处理复杂资格标准以及提取复杂匹配逻辑的研究25。刘等。探索了GPT-4在放射域特异性用例中的性能26。Nori等。进一步发现,具有高级及时工程的通用域LLM可以在不进行微调的情况下实现最高准确性27。最近的评论还详细总结了相关研究28,,,,29,,,,30。
这些结果证明了在BionLP应用程序中使用LLM的潜力,尤其是当可以使用最小的手动策划黄金标准数据时,并且不需要为每项新任务进行微调或重新训练。在生物医学领域中,主要挑战是标记数据集的可用性有限,该数据集的规模明显低于一般域中的规模(例如,生物医学句子相似性数据集仅具有100个标记的实例,总计31)32,,,,33。这挑战了微调方法,因为(1)在有限标记的数据集上进行微调的模型可能无法推广,并且(2)对具有更大尺寸的模型进行微调变得更具挑战性。
受早期实验的促进,系统地评估LLMS在BIONLP任务中的有效性并理解其对Bionlp方法开发和下游用户的影响很重要。桌子 1在这种情况下,提供了代表性研究的详细比较。虽然我们的主要重点是生物医学领域,特别是使用生物医学文献对LLM的评估,但我们还在临床领域中包括了两项代表性研究(使用临床记录评估LLMS)供参考。有几个主要局限性。首先,大多数评估研究主要评估了GPT-3或GPT-3.5,这些研究可能无法提供来自不同类别的代表性LLM。例如,很少有研究评估了更先进的闭合源LLM,例如GPT-4,LLM代表,来自Llama等一般领域22和生物医学领域特异性LLM,例如PMC-llama34。其次,现有研究主要评估了固定黄金标准的提取任务。这些研究中很少有人评估生成任务,例如文本摘要和文本简化,而金标准是自由文本的。可以说,现有的变压器模型在提取任务中表现出令人满意的性能,而生成任务在达到相似水平的水平方面仍然是一个挑战。因此,必须评估BIONLP生成任务中LLM的有效性,以检查它们是否可以补充现有模型。第三,大多数现有研究仅报告了定量评估,例如F1得分,对定性评估的重视程度有限。但是,进行定性评估(例如,评估LLM生成的文本的质量并分类不一致或幻觉的响应)以了解LLM对生物医学领域下游应用的错误和影响比单纯的定量计量更为关键。例如,对LLM的研究发现,人类判断与自动措施(例如Rouge-l)之间的相关相关性相对较低,通常应用于临床领域中的文本摘要任务35。最后,值得注意的是,几项研究没有提供公众访问其相关数据或代码的访问。例如,很少有研究提示或精选的示例,以进行几次学习。这阻碍了可重复性,并且在使用相同的设置进行公平比较的新LLMS评估新LLM方面也提出了挑战。
在这项研究中,我们对BIONLP应用中的LLM进行了全面评估,以检查其巨大潜力及其局限性和错误。我们的研究有三个主要贡献。
首先,我们对四个代表性LLM进行了全面评估:GPT-3.5和GPT-4(封闭源LLMS的代表),Llama 2(来自开源LLMS的代表)和PMC Llama(来自生物医学领域特异性LLMS的代表性)。We evaluated them on 12 BioNLP datasets across six applications: (1) named entity recognition, which extracts biological entities of interest from free-text, (2) relation extraction, which identifies relations among entities, (3) multi-label document classification, which categorizes documents into broad categories, (4) question answering, which provides answers to medical questions, (5) text summarization, which produces a coherent summary of an input文本和(6)文本简化,它生成输入文本的可理解内容。在四个设置下评估了这些模型:零射击,静态射击,动态k-nearest几乎没有射击和微调。我们将这些模型与使用微调的,特定于域的BERT或BART模型的最新模型(SOTA)方法进行了比较。BERT和BART模型在Bionlp研究中都有良好的发展。
我们的结果表明,在大多数BIONLP任务中,SOTA微调方法的表现优于零和少数LLM。这些方法的宏观平均水平比12个基准(0.65 vs. 0.51)的最佳零和少数LLM性能高约15%,信息提取任务(例如,关系提取(0.79 vs. 0.33))高出40%以上。但是,诸如GPT-3.5和GPT-4之类的封闭源LLM在与推理相关的任务(例如医疗问题回答)中表现出更好的零和很少的性能,它们在何处优于SOTA微调方法。此外,他们在与生成相关的任务(例如文本摘要和简化)中表现出比较低但合理的性能,表现出竞争性的准确性和可读性,并在语义理解任务(例如文档级分类)中显示出潜力。在LLMS中,GPT-4的总体表现最高,尤其是由于其出色的推理能力。但是,它具有权衡,比GPT-3.5贵6至100倍。相比之下,诸如Llama 2之类的开源LLM并未显示出强大的零和很少的性能 - 它们仍然需要微调来弥合Bionlp应用程序的性能差距。
其次,我们对来自LLM的数十万个样本产出进行了彻底的手动验证。对于固定黄金标准的提取和分类任务(例如,关系提取和多标签文档分类),我们检查了(1)丢失的输出,当LLM无法提供所需的输出时,(2)当LLMS的输出不一致时,当LLMS用于类似实例的不同输出时,以及(3)幻觉输出,(3)幻觉输出,(3)可能会在幻觉中误解,并且可能会在幻觉中无法解决用户的用户,并且可能会在用户限制方面误解,并且可能会在用户限制中误解。36。对于文本摘要任务,两名医疗保健专业人员进行了手动评估,以评估准确性,完整性和可读性。结果表明,普遍的丢失,不一致和幻觉输出的案例,尤其是在零射击设置下的美洲驼。例如,多标签文档分类数据集有102多个幻觉案例(占总测试实例的32%)和69个不一致的情况(22%)。
最后,我们为下游用户提供了最佳实践中使用LLMS在BionLP应用程序中的建议。我们还注意到了两个开放问题。首先,BIONLP中的当前数据和评估范例是根据监督方法量身定制的,对LLM可能不公平。例如,结果表明,用于文本摘要的自动指标可能与手动评估不符。同样,在生物医学领域中,专门针对LLMS Excel(例如推理)的数据集受到限制。重新访问Bionlp中的数据和评估范例是最大化LLM在BIONLP应用程序中的好处的关键。其次,解决错误,丢失信息和不一致的问题对于最大程度地降低了生物医学和临床应用中与LLM相关的风险至关重要。我们强烈鼓励社区努力找到更好的解决方案来减轻这些问题。
我们认为,这项研究的发现将对Bionlp下游用户有益,也将有助于进一步提高BIONLP应用程序中LLM的性能。已建立的基准和基线性能可以作为评估生物医学领域中新LLM的基础。为了确保可重复性并促进基准测试,我们已经可以通过公开访问的相关数据,模型和结果https://doi.org/10.5281/Zenodo.1402550037。
结果
定量评估
桌子 2说明了12个数据集的主要评估度量结果及其在零/少数射击(静态单次和五射)和微调设置下的LLM的宏观大小。特定数据集的结果与其他研究独立报道的结果一致,例如GPT-3.5零射击和GPT-4零拍摄的MEDQA的准确性为0.4462和0.7471(我们的研究中分别为0.4988和0.7156)38。同样,分别报道了GPT-3.5零射门的HOC和LITCOVID的MICRO-F1和0.6720(分别为0.6605和0.6707)(分别为0.6605和0.6707)39。还报道了PubMedQA上0.7790的精度,用于微调PMC Llama 13B(合并多个问题回答用于微调的数据集)34;我们的研究还仅使用PubMedQA训练集报道了类似的0.7680精度。我们进一步总结了补充信息的详细结果 S2定量评估结果,包括S2.2中的次级度量结果,s2.3中的性能平均值,方差和置信区间,统计测试在S2.4中结果和动态k-nearest最少的少数射击结果S2.5。
Sota vs. LLMS。表中提供了SOTA微调方法的结果 2。回想一下,SOTA方法使用了微调(特定领域)的语言模型。对于提取和分类任务,SOTA接近微调的生物医学领域特异性BERT模型,例如Biobert和PubMedbert。对于文本摘要和简化任务,SOTA接近微调的BART模型。
如表中所示 2,SOTA微调方法在12个数据集的宏平均值为0.6536,而最佳LLM对应物分别为0.4561、0.4750、0.4862和0.5131,在零发射,一次性,五杆和精细设置下。在12个数据集中的10个中,它的表现优于LLM的零和几射击。它具有更高的性能,尤其是在信息提取任务中。例如,对于NCBI疾病,SOTA方法达到了实体级别的F1得分为0.9090,而在零和一次性设置下,LLMS(GPT-4)的最佳结果降低了30%(0.5988)。在微调设置下,LLM的性能更接近,Llama 2 13B达到了实体级别的F1得分为0.8682,但仍然较低。值得注意的是,SOTA微调方法是非常强大的基线 - 与基础模型相比,它们比简单的微调要复杂得多。继续以NCBI疾病为例,SOTA微调方法产生了大规模的弱标记示例,并使用对比度学习来学习一般表示。
相比之下,LLM的表现优于有关回答的SOTA微调方法。对于MEDQA,SOTA方法的精度为0.4195。在零射击设置下的GPT-4在绝对差的精度(0.7156)的精度上提高了近30%,而GPT-3.5在零弹位设置下的精度也提高了约8%(0.4988)。对于PubMedQA,SOTA方法的精度为0.7340。GPT-4在一次性设置下的精度具有相似的精度(0.7100),并且具有更高的精度,并以更多的射击(在五杆设置下为0.7580),正如我们将稍后显示的那样。在微调设置(分别为0.8040和0.7680)下,Llama 2 13B和PMC Llama 13B的精度也更高。在这种情况下,GPT-3.5在SOTA方法上没有达到更高的精度,但是在五弹性设置下它已经具有竞争精度(0.6950)。
LLMS之间的比较。在LLMS中,在零/少数射击设置下进行比较,结果表明GPT-4的性能始终具有最高的性能。在零射击设置下,GPT-4的宏观平均水平为0.4561,比GPT-3.5(0.3814)高约7%,几乎是Llama 2 13B(0.2362)的两倍。它在12个数据集中的9个中达到了最高的性能,其性能也占其余三个数据集最佳结果的3%以内。一次性和五弹性设置显示出非常相似的模式。
此外,在零和一击设置下,Llama2 13B的性能大大低于GPT-3.5(低15%,低15%)和GPT-4(降低22%,降低22%,降低22%,低17%)。与最佳LLM结果相比,在特定数据集中的性能低达六倍。例如,在零弹性设置下为0.1286 vs. 0.7109。这些结果表明,Llama2 13B仍然需要微调才能达到相似的性能并弥合性能差距。微调将骆驼2 13B的宏观平均含量从0.2837提高到0.5131。值得注意的是,其在微调设置下的性能略高于GPT-4的零和少量性能。微调美洲驼2 13B通常在所有任务中都提高了其性能,除了文本摘要和文本简化。其性能限制的一个关键原因是,数据集的输入上下文要比其允许的输入令牌(4096)更长,因此在这种情况下进行微调无济于事。该观察结果还激发了进一步的研究工作,以扩展LLMS上下文窗口40,,,,41。在微调环境下,结果还表明,作为持续预处理的生物医学领域特异性LLM,PMC Llama 13B并没有达到总体上比Llama 2 13B更高的性能。微调的美洲驼2 13B的性能要比12个数据集中的10个PMC Llama 13B更好。
如前所述,我们再现了PMC Llama研究中报道的类似结果34。例如,它报告了PubMedQA上0.7790的准确性,并通过微调多个问题回答数据集。仅在PubMedQA数据集上微调PMC Llama 13B时,我们的精度非常相似,为0.7680。但是,我们还发现,使用完全相同的设置对Llama 2 13B进行微调导致更好或至少相似的性能。
几乎没有成本分析
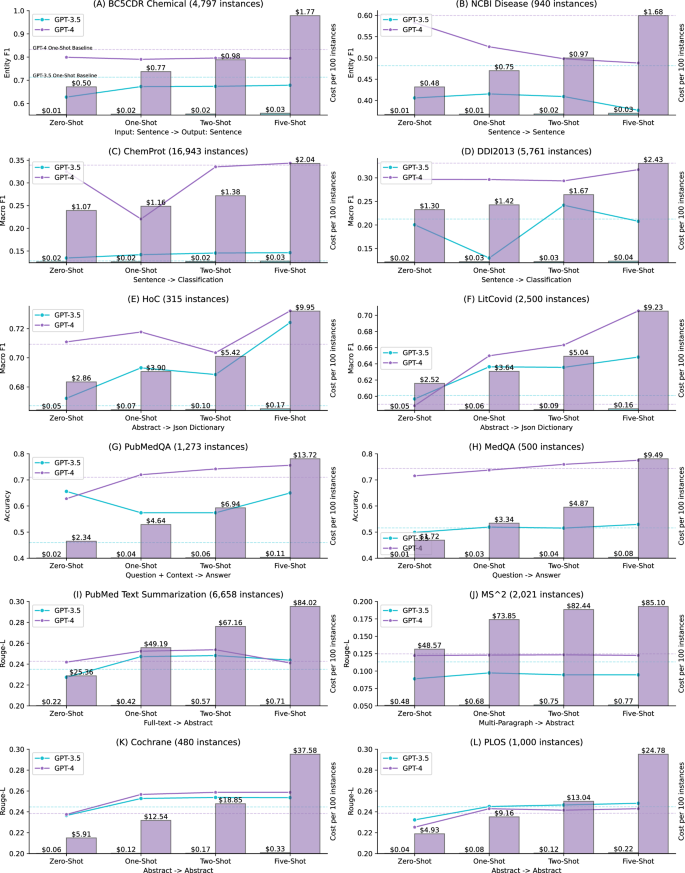
图 1进一步说明了动态k-nearest最少的射击的性能以及相关的成本随射击数量的增加而相关的成本。详细的结果还提供了补充信息 S2。对于一个,二和五的k值进行了动态k-nearest射击。为了进行比较,我们还提供了该图中的零射击和静态的一击性能。结果表明,动态k-nearest最少的射击最有效,对于多标签文档分类和问题答案。例如,对于litcovid数据集,GPT-4在静态单发设置下的宏F1为0.5901;相比之下,在动态最终射击下,其宏F1的宏F1为0.6500,并进一步增加到0.7055,射击了五个射击。类似地,GPT-3.5也表现出改进,其宏F1在静态单枪设置下为0.6009,而动态一击和五杆分别为0.6364和0.6484。对于问题回答,对于多标签文档分类而言,改进不如多标签文档分类,但总体趋势显示出稳定的增长,尤其是考虑到GPT-4的性能已经比零射击的SOTA方法相似或更高。例如,其在PubMedQA上的准确性为0.71,具有静态镜头。在动态的一击和五射击下,精度分别提高到0.72和0.75。
数据和方法部分总结了几次射击和成本分析的详细方法。动态k-nearest几射击涉及选择k最近的训练实例作为每个测试实例的示例。此外,静态单发的性能(使用相同的单发示例为每个测试实例)显示为虚线的水平线以进行比较。补充信息还提供了数字的详细性能 S2。
相比之下,结果表明,动态k-nearest最少的射击对其他任务的有效性较差。例如,动态的单发性能低于两个命名实体识别数据集上两个GPT模型的静态单发性能,并且通过增加动态镜头的数量也无济于事。在关系提取中也观察到类似的发现。对于文本摘要和文本简化任务,在两个数据集中,动态的K-Neart最少的射击性能略高,但是通常,它与静态的一击性能非常相似。此外,结果还表明,增加镜头的数量并不一定会提高性能。例如,具有动态五射线的GPT-4在12个数据集中的八个中没有最高的性能。在其他研究中也报道了类似的发现,其中GPT-3.5具有五次学习的表现低于自然语言推理任务的零摄像学习39。
图 1进一步比较了使用GPT-3.5和GPT-4的每100个实例的成本。成本是根据单价价格的输入和输出令牌数量计算的。我们将GPT-4-0613用于提取任务,而GPT-4-32K-0613用于生成任务,因为输入和输出上下文更长的时间更长,尤其是更多的镜头。GPT-4通常表现出最高的性能,如两个表所示 2和图 1;但是,成本分析结果也表明了一个明确的权衡,GPT-4的昂贵60至100倍。对于提取性和分类任务,五击的每100个实例的实际成本从句子级别的输入约2美元到抽象级别输入的$ 10左右。这一成本是GPT-3.5的60至70倍,句子级输入的价格约为0.03美元,抽象级输入的句子约为0.16美元。对于生成任务,成本差异更为明显,比例比例更高100倍或更昂贵。原因之一是GPT-432âK的单位价格更高,文本摘要之类的任务涉及更长的输入和输出令牌。以PubMed文本摘要数据集为例,GPT-4的价格为$ 84.02每100个实例,五次摄,总计约5600美元,用于推断整个测试集。相比之下,GPT-3仅花费五发情况每100个实例0.71美元,整个测试集的总计约为48美元。
基于绩效和成本结果,它表明成本差不一定会扩展到绩效差异,除了回答任务。GPT-4在提问任务中的精度比GPT-3.5高20%至30%,并且高于SOTA方法。对于其他任务,性能差异要小得多,成本明显更高。例如,在两个文本简化任务上的GPT-4的性能在GPT-3.5的2%以内,但实际成本高于100倍以上。
定性评估
命名实体识别的错误分析
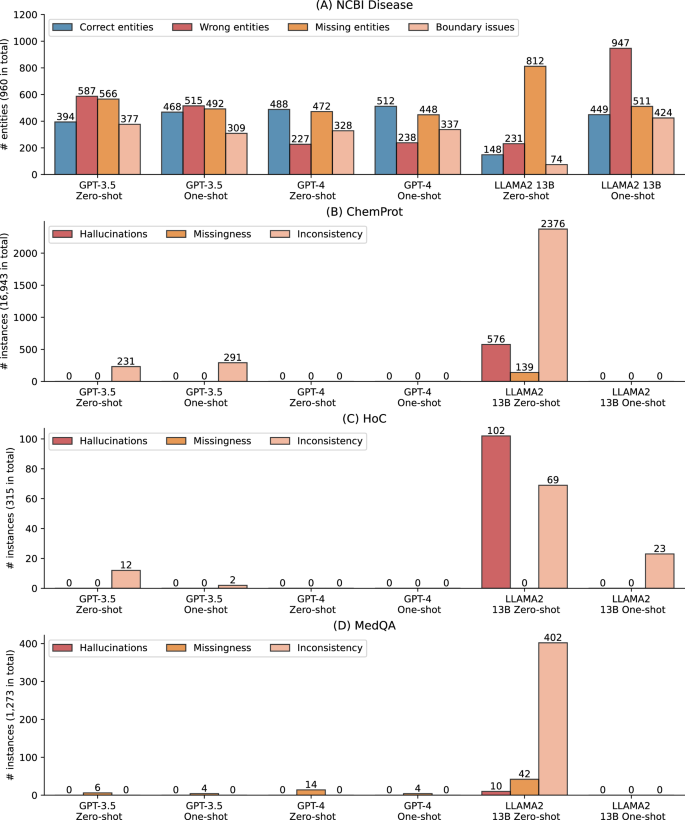
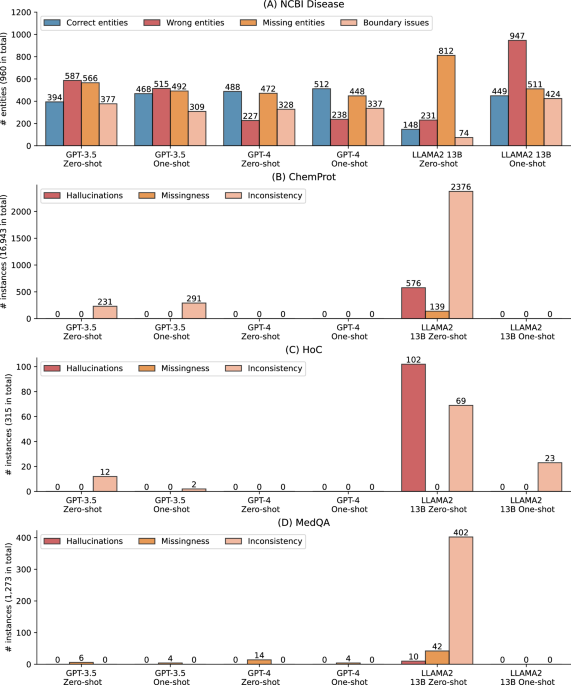
图 2a进一步显示了对命名实体识别基准NCBI疾病的错误分析,其中零和几乎没有射击设置下的LLM的性能大大低于SOTA结果(例如,Llama 2 13B零发性能低70%)。回想一下,指定的实体识别提取物从自由文本中提取了实体,基准测试评估了这些提取实体的准确性。我们检查了完整测试集中的所有预测,并将其分为四种类型:(1)正确的实体,其中预测的实体在文本跨度和实体类型中都是正确的,(2)错误的实体,(2)在其中预测的实体是不正确的,(3)缺失实体,在其中预测的跨度标准,以及(4)跨度标准,以及(4)跨度标准,以及(4)跨度,以及(4)范围的范围,以及(4)范围的范围,以及(4)范围的范围,以及(4)范围。 2a。结果表明,LLMS总共可以预测960个中多达512个实体,从而解释了较低的F1得分。由于SOTA模型尚未公开,我们使用独立研究的NCBI疾病使用了替代的微调生物模型(https://huggingface.co/ugaray96/biobert_ncbi_disease_ner),该实体级别的F1得分为0.8920用于比较。它正确预测了960条中的863个实体。错误的实体,缺失实体和边界问题分别为111、97和269。
另外,图 2aalso shows that GPT-4 had the lowest number of wrong entities, whereas other categories have a similar prevalence to GPT-3.5, which explains its higher F1-score overall.Furthermore, providing one shot did not alter the errors for GPT-3.5 and GPT-4 compared to their zero-shot settings, but it dramatically changed the results for LLaMA 2 13B.Under one-shot, LLaMA 2 13B had 449 correctly predicted entities, compared to 148 under zero-shot.Additionally, its missing entities also reduced from 812 to 511 with one-shot, but it also had a trade-off of more boundary issues and wrong entities.
Evaluations on inconsistencies, missing information, and hallucinations
Figure 2B–Dpresent the qualitative evaluation results on ChemProt, HoC, and MedQA, respectively.Recall that we categorized inconsistencies, missing information, and hallucinations on the tasks where the gold standard is a fixed classification type or a multiple-choice option.Table 3also provides detailed examples.The findings show prevalent inconsistent, missing, or hallucinated responses, particularly in LLaMA 2 13B zero-shot responses.For instance, it exhibited 506 hallucinated responses (~3% out of the total 16,943 instances) and 2376 inconsistent responses (14%) for ChemProt.In the case of HoC, there were 102 (32%) hallucinated responses and 69 (22%) inconsistent responses.Similarly, for MedQA, there were 402 (32%) inconsistent responses.In comparison, GPT-3.5 and GPT-4 exhibited substantially fewer cases.GPT-3.5 showed a small number of inconsistent responses for ChemProt and HoC, and a few missing responses for MedQA.On the other hand, GPT-4 did not exhibit any such cases for ChemProt and HoC, while displaying a few missing responses for MedQA.
It is worth noting that inconsistent responses do not necessarily imply that they fail to address the prompts;rather, the responses answer the prompt but in different formats.In contrast, hallucinated cases do not address the prompts and may repeat the prompts or contain irrelevant information.All such instances pose challenges for automatic extraction or postprocessing and may require manual review.As a potential solution, we observed that adding just one shot could significantly reduce such cases, especially for LLaMA 2 13B, which exhibited prevalent instances in zero-shot.As illustrated in Fig. 2b, LLaMA 2 13B one-shot dramatically reduced these cases in ChemProt and MedQA.Similarly, its hallucinated responses decreased from 102 to 0, and inconsistent cases decreased from 69 to 23 in HoC with one-shot.Another solution is fine-tuning, which we did not find any such cases during the manual examination, albeit with a trade-off of computational resources.
Evaluations on accuracy, completeness, and readability
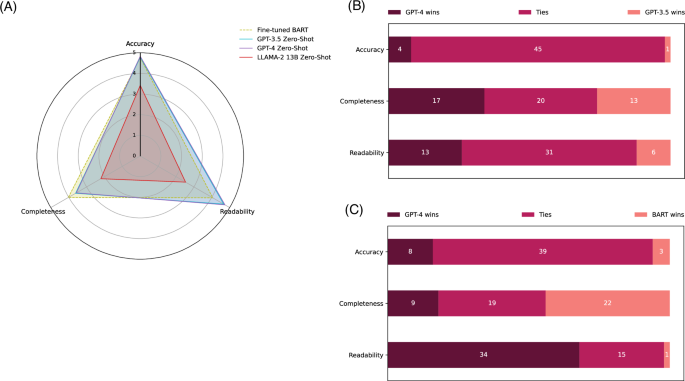
Figure 3presents the qualitative evaluation results on the PubMed Text Summarization dataset.In Fig. 3a, the overall results in accuracy, completeness, and readability for the four models on 50 random samples are depicted.The evaluation results in digits are further demonstrated in Table 4for complementary.Detailed results with statistical analysis and examples are available in Supplementary Information S3。The fine-tuned BART model used in the SOTA approach42, serving as the baseline, achieved an accuracy of 4.76 (out of 5), a completeness of 4.02, and a readability of 4.05.In contrast, both GPT-3.5 and GPT-4 demonstrated similar and slightly higher accuracy (4.79 and 4.83, respectively) and statistically significantly higher readability than the fine-tuned BART model (4.66 and 4.73), but statistically significantly lower completeness (3.61 and 3.57) under the zero-shot setting.The LLaMA 2 13B zero-shot performance is substantially lower in all three aspects.
一个The overall results of the fine-tuned BART, GPT-3.5 zero-shot, GPT-4 zero-shot, and LLaMA 2 zero-shot models on a scale of 1 to 5, based on random 50 testing instances from the PubMed Text Summarization dataset.b和cdisplay the number of winning, tying, and losing cases when comparing GPT-4 zero-shot to GPT-3.5 zero-shot and GPT-4 zero-shot to the fine-tuned BART model, respectively.Table 4shows the results in digits for complementary.Detailed results, including statistical tests and examples, are provided in Supplementary Information S3。
3bfurther compares GPT-4 to GPT-3.5 and the fine-tuned BART model in detail.In the comparison between GPT-4 and GPT-3.5, GPT-4 had a slightly higher number of winning cases in the three aspects (4 winning cases vs. 1 losing case for accuracy, 17 vs. 13 for completeness, and 13 vs. 6 for readability).Most of the cases resulted in a tie.When comparing GPT-4 to the fine-tuned BART model, GPT-4 had significantly more winning cases for readability (34 vs. 1) with much fewer winning cases for completeness (9 vs. 22).
讨论
First, the SOTA fine-tuning approaches outperformed zero- and few-shot performance of LLMs in most of BioNLP applications.As demonstrated in Table 2, it had the best performance in 10 out of the 12 benchmarks.In particular, it outperformed zero- and few-shot LLMs by a large margin in information extraction and classification tasks such as named entity recognition and relation extraction, which is consistent to the existing studies43,,,,44。In contrast to, other tasks such as medical question answering, named entity recognition, and relation extraction require limited reasoning and extract information directly from inputs at the sentence-level.Zero- and few-shot learning may not be appropriate or sufficient for these conditions.For those tasks, arguably, fine-tuned biomedical domain-specific language models are still the first choice and have already set a high bar, according to the literature32。
In addition, closed-source LLMs such as GPT-3.5 and GPT-4 demonstrated reasonable zero- and few-shot capabilities for three BioNLP tasks.The most promising task that outperformed the SOTA fine-tuning approaches is medical question answering, which involves reasoning45。As shown in Table 2and Fig. 1, GPT-4 already outperformed previous fine-tuned SOTA approaches in MedQA and PubMedQA with zero- or few-shot learning.This is also supported by the existing studies on medical question answering38,,,,46。The second potential use case is text summarization and simplification.As shown in Table 2, those tasks are still less favored by the automatic evaluation measures;however, manual evaluation results show both GPT-3.5 and GPT-4 had higher readability and competitive accuracy compared to the SOTA fine-tuning approaches.Other studies reported similar findings regarding the low correlation between automatic and manual evaluations35,,,,47。The third possible use case – though still underperformed by previous fine-tuned SOTA approaches – document-level classification, which involves semantic understanding.As shown in Fig. 1, GPT-4 achieved over a 0.7 F1-score with dynamic K-nearest shot for both multi-label document-level classification benchmarks.
In addition to closed-source LLMs, open-source LLMs such as LLaMA 2 do not demonstrate strong zero- and few-shot capabilities.While there are other open-source LLMs available, LLaMA 2 remains as a strong representative48。Results in Table 1suggest that its overall zero-shot performance is 15% and 22% lower than that of GPT-3.5 and GPT-4, respectively, and up to 60% lower in specific BioNLP tasks.Not only does it exhibit suboptimal performance, but the results in Fig. 2also demonstrate that its zero-shot responses frequently contain inconsistencies, missing elements, and hallucinations, accounting for up to 30% of the full testing set instances.Therefore, fine-tuning open-source LLMs for BioNLP tasks is still necessary to bridge the gap.Only through fine-tuning LLaMA 2, its overall performance is slightly higher than the one-shot GPT-4 (4%).However, it is worth noting that the model sizes of LLaMA 2 and PMC LLaMA are significantly smaller than those of GPT-3.5 and GPT-4, making it challenging to evaluate them on the same level.Additionally, open-source LLMs have the advantage of continued development and local deployment.
Another primary finding on open-source LLMs is that the results do not indicate significant performance improvement from continuously biomedical pre-trained LLMs (PMC LLaMA 13B vs. LLaMA 2 13B).As mentioned, our study reproduced similar results reported in PMC LLaMA 13B;however, we also found that directly fine-tuning LLaMA 2 yielded better or at least similar performance—and this is consistent across all 12 benchmarks.In the biomedical domain, representative foundation LLMs such as PMC LLaMA used 32 A100 GPUs34, and Meditron used 128 A100 GPUs to continuously pretrain from LLaMA or LLaMA 249。Our evaluation did not find significant performance improvement for PMC LLaMA;the Meditron study also only reported ~3% improvement itself and only evaluated on question answering datasets.At a minimum, the results suggest the need for a more effective and sustainable approach to developing biomedical domain-specific LLMs.
The automatic metrics for text summarization and simplification tasks may not align with manual evaluations.As the quantitative results on text summarization and generation demonstrated, commonly used automatic evaluations such as Rouge, BERT, and BART scores consistently favored the fine-tuned BART’s generated text, while manual evaluations show different results, indicating that GPT-3.5 and GPT-4 had competitive accuracy and much higher readability even under the zero-shot setting.Existing studies also reported that the automatic measures on LLM-generated text may not correlate to human preference35,,,,47。The MS^2 benchmark used in the study also discussed the limitation of automatic measures, specifically for text summarization50。Additionally, the results highlight that completeness is a primary limitation when adapting GPT models to biomedical text generation tasks despite its competitive accuracy and readability scores.
Last, our evaluation on both performance and cost demonstrates a clear trade-off when using LLMs in practice.GPT-4 had the overall best performance in the 12 benchmarks, with an 8% improvement over GPT-3.5 but also at a higher cost (60 to 100 times higher than GPT-3.5).Notably, GPT-4 showed significantly higher performance, particularly in question-answering tasks that involve reasoning, such as over 20% improvement in MedQA compared to GPT-3.5.This observation is consistent with findings from other studies27,,,,38。Note that newer versions of GPT-4, such as GPT-4 Turbo, may further reduce the cost of using GPT-4.
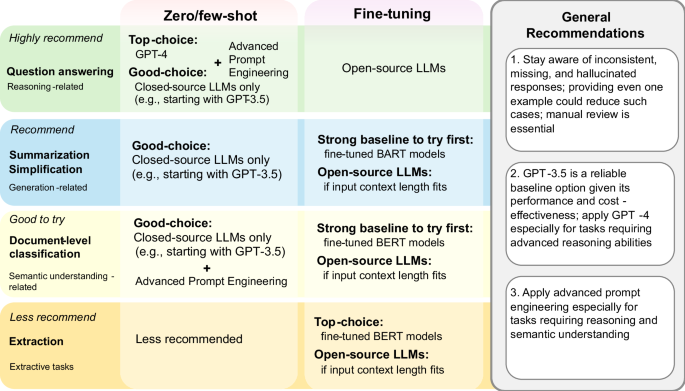
These findings lead to recommendations for downstream users to apply LLMs in BioNLP applications, summarized in Fig. 4。It provides suggestions on which BioNLP applications are recommended (or not) for LLMs, categorized by conditions (e.g., the zero/few-shot setting when computational resources are limited) and additional tips (e.g., when advanced prompt engineering is more effective).
We also recognize the following two open problems and encourage a community effort for better usage of LLMs in BioNLP applications.
Adapting both data and evaluation paradigms is essential to maximize the benefits of LLMs in BioNLP applications.Arguably, the current datasets and evaluation settings in BioNLP are tailored to supervised (fine-tuning) methods and is not fair for LLMs.Those issues challenge the direct comparison between the fine-tuned biomedical domain-specific language models and zero/few shot of LLMs.The datasets for the tasks where LLMs excel are also limited in the biomedical domain.Further, the manual measures on biomedical text summarization also showed different results than that of all three automatic measures.These collectively suggest the current BioNLP evaluation frameworks have limitations when they are applied to LLMs35,,,,51。They may not be able to accurately assess the full benefits of LLMs in biomedical applications, calling for the development of new evaluation datasets and methods for LLMs in bioNLP tasks.
Addressing inconsistencies, missingness, and hallucinations produced by LLMs is critical.The prevalence of inconsistencies, missingness, and hallucinations generated by LLMs is of concern, and we argue that they must be addressed for deployment.Our results demonstrate that providing just one shot could significantly reduce the occurrence of such issues, offering a simple solution.However, thorough examination in real-world scenario validations is still necessary.Additionally, more advanced approaches for validating LLMs’ responses are expected for further improvement of their reliability and usability47。
This study also has several limitations that should be acknowledged.While this study examined strong LLM representatives from each category (closed-source, open-source, and biomedical domain-specific), it is important to note that there are other LLMs, such as BARD52and Mistral53, that have demonstrated strong performance in the literature.Additionally, while we investigated zero-shot, one-shot, dynamic K-nearest few-shot, and fine-tuning techniques, each of them has variations, and there are also new approaches54。Given the rapidly growing nature of this area, our study cannot cover all of them.Instead, our aim is to establish baseline performance on the main BioNLP applications using commonly used LLMs and methods as representatives, and to make the datasets, methods, codes, and results publicly available.This enables downstream users to understand when and how to apply LLMs in their own use cases and to compare new LLMs and associated methods on the same benchmarks.In the future, we also plan to assess LLMs in real-world scenarios in the biomedical domain to further broaden the scope of the study.
方法
Evaluation tasks, datasets, and metrics
Table 5presents a summary of the evaluation tasks, datasets, and metrics.We benchmarked the models on the full testing sets of the twelve datasets from six BioNLP applications, which are BC5CDR-chemical and NCBI-disease for Named Entity Recognition, ChemProt and DDI2013 for relation extraction, HoC and LitCovid for multi-label document classification, and MedQA and PubMedQA for question answering, PubMed Text Summarization and MS^2 for text summarization, and Cochrane PLS and PLOS Text Simplification for text simplification.These datasets have been widely used in benchmarking biomedical text mining challenges55,,,,56,,,,57and evaluating biomedical language models9,,,,10,,,,11,,,,16。The datasets are also available in the repository.We evaluated the datasets using the official evaluation metrics provided by the original dataset description papers, as well as commonly used metrics for method development or applications with the datasets, as documented in Table 5。Note that it is challenging to have a single one-size-fits-all metric, and some datasets and related studies used multiple evaluation metrics.Therefore, we also adopted secondary metrics for additional evaluations.A detailed description is below.
Named entity recognition.Named entity recognition is a task that involves identifying entities of interest from free text.The biomedical entities can be described in various ways, and resolving the ambiguities is crucial58。Named entity recognition is typically a sequence labeling task, where each token is classified into a specific entity type.BC5CDR-chemical59and NCBI-disease60are manually annotated named entity recognition datasets for chemicals and diseases mentioned in biomedical literature, respectively.The exact match (that is, the predicted tokens must have the same text spans as the gold standard) F1-score was used to quantify the model performance.
Relation extraction.Relation extraction involves identifying the relationships between entities, which is important for drug repurposing and knowledge discovery61。Relation extraction is typically a multi-class classification problem, where a sentence or passage is given with identified entities and the goal is to classify the relation type between them.ChemProt55and DDI201362are manually curated relation extraction datasets for protein-protein interactions and drug-drug interactions from biomedical literature, respectively.Macro and micro F1-scores were used to quantify the model performance.
Multi-label document classification.Multi-label document classification identifies semantic categories at the document-level.The semantic categories are effective for grasping the main topics and searching for relevant literature in the biomedical domain63。Unlike multi-class classification, which assigns only one label to an instance, multi-label classification can assign up to N labels to an instance.HoC64and LitCovid56are manually annotated multi-label document classification datasets for hallmarks of cancer (10 labels) and COVID-19 topics (7 labels), respectively.Macro and Micro F1 scores were used as the primary and secondary evaluation metrics, respectively.
Question answering.Question answering evaluates the knowledge and reasoning capabilities of a system in answering a given biomedical question with or without associated contexts45。Biomedical QA datasets such as MedQA and PubMedQA have been widely used in the evaluation of language models65。The MedQA dataset is collected from questions in the United States Medical License Examination (USMLE), where each instance contains a question (usually a patient description) and five answer choices (e.g., five potential diagnoses)66。The PubMedQA dataset includes biomedical research questions from PubMed, and the task is to use yes, no, or maybe to answer these questions with the corresponding abstracts67。Accuracy and macro F1-score are used as the primary and secondary evaluation metrics, respectively.
Text summarization.Text summarization produces a concise and coherent summary of a longer documents or multiple documents while preserving its essential content.We used two primary biomedical text summarization datasets: the PubMed text summarization benchmark68and MS^250。The PubMed text summarization benchmark focuses on single document summarization where the input is a full PubMed article, and the gold standard output is its abstract.M2^2 in contrast, focuses on multi-document summarization where the input is a collection of PubMed articles, and the gold standard output is the abstract of a systematic review study that cites those articles.Both benchmarks used the ROUGE-L score as the primary evaluation metric;BERT score and BART score were used as secondary evaluation metrics.
Text simplification.Text simplification rephrases complex texts into simpler language while maintaining the original meaning, making the information more accessible to a broader audience.We used two primary biomedical text simplification datasets: Cochrane PLS69and the PLOS text simplification benchmark70。Cochrane PLS consists of the medical documents from the Cochrane Database of Systematic Reviews and the corresponding plain-language summary (PLS) written by the authors.The PLOS text simplification benchmark consists of articles from PLOS journals and the corresponding technical summary and PLS written by the authors.The ROUGE-L score was used as the primary evaluation metric.Flesch-Kincaid Grade Level (FKGL) and Dale-Chall Readability Score (DCRS), two commonly used evaluation metrics on readability71were used as the secondary evaluation metrics.
基线
For each dataset, we reported the reported SOTA fine-tuning result before the rise of LLMs as the baseline.The SOTA approaches involved fine-tuning (domain-specific) language models such as PubMedBERT16, BioBERT9, or BART72as the backbone.The fine-tuning still requires scalable manually labeled instances, which is challenging in the biomedical domain32。In contrast, LLMs may have the advantage when minimal manually labeled instances are available, and they do not require fine-tuning or retraining for every new task through zero/few-shot learning.Therefore, we used the existing SOTA results achieved by the fine-tuning approaches to quantify the benefits and challenges of LLMs in BioNLP applications.
大型语言模型
Representative LLMs and their versions.Both GPT-3.5 and GPT-4 have been regularly updated.For reproducibility, we used the snapshots gpt-3.5-turbo-16k-0613 and gpt-4-0613 for extractive tasks, and gpt-4-32k-0613 for generative tasks, considering their input and output token sizes.Regarding LLaMA 2, it is available in 7B, 13B, and 70B versions.We evaluated LLaMA 2 13B based on the computational resources required for fine-tuning, which is arguably the most common scenario applicable to BioNLP downstream applications.For PMC LLaMA, both 7B and 13B versions are available.Similarly, we used PMC LLaMA 13B, specifically evaluating it under the fine-tuning setting – the same setting used in its original study34。In the original study, PMC LLaMA was only evaluated on medical question answering tasks, combining multiple question answering datasets for fine-tuning.In our case, we fine-tuned each dataset separately and reported the results individually.
Prompts.To date, prompt design remains an open research problem73,,,,74,,,,75。We developed a prompt template that can be used across different tasks based on existing literature74,,,,75,,,,76,,,,77。An annotated prompt example is provided in Supplementary Information S1Prompt engineering, and we have made all the prompts publicly available in the repository.The prompt template contains (1) task descriptions (e.g., classifying relations), (2) input specifications (e.g., a sentence with labeled entities), (3) output specifications (e.g., the relation type), (4) task guidance (e.g., detailed descriptions or documentations on relation types), and (5) example demonstrations if examples from training sets are provided.This approach aligns with previous studies in the biomedical domain, which have demonstrated that incorporating task guidance into the prompt leads to improved performance74,,,,76and was also employed and evaluated in our previous study, specifically focusing on named entity recognition77。We also adapted the SOTA example selection approach in the biomedical domain described below27。
Zero-shot and static few-shot.We comparatively evaluated the zero-shot, one-shot, and five-shot learning performances.Only a few studies have made the selected examples available.For reproducibility and benchmarking, we first randomly selected the required number of examples in training sets, used the same selected examples for few-shot learning, and made the selected examples publicly available.
Dynamic K-nearest few-shot.In addition to zero- or static few-shot learning where fixed instructions are used for each instance, we further evaluated the LLMs under a dynamic few-shot learning setting.The dynamic few-shot learning is based on the MedPrompt approach, the SOTA method that demonstrated robust performance in medical question answering tasks without fine-tuning27。The essence is to use K training instances that are most similar to the test instance as the selected examples.We denote this setting as dynamic K-nearest few-shot, as the prompts for different test instances differ.Specifically, for each dataset, we used the SOTA text embedding model text-embedding-ada-00254to encode the instances and used cosine similarity as the metric for finding similar training instances to a testing instance.We tested dynamic K-nearest few-shot prompts with K equals to one, two, and five.
Parameters for prompt engineering.For zero-, one-, and few-shot approaches, we used a temperature parameter of 0 to minimize variance for both GPT and LLaMA-based models.Additionally, for LLaMA models, we maintained other parameters unchanged, set the maximum number of generated tokens per task, and truncated the instances due to the input length limit for the five-shot setting.Further details are provided in Supplementary Information S1Prompt engineering, and the related codes are available in the repository.
微调。We further conducted instruction fine-tuning on LLaMA 2 13B and PMC-LLaMA 13B.For each dataset, we fine-tuned LLaMA 2 13B and PMC- LLaMA 13B using its training set.The goal of instruction fine-tuning is defined by the objective function:\({\arg }{\max }_{\theta }{\sum}_{\left({x}^{i},{y}^{i}\right)\epsilon (X,Y)}{logp}({y}^{i}|{x}^{i};\theta )\), 在哪里\({x}^{i}\)represents the input instruction,\({y}^{i}\)is the ground truth response, and\(\theta\)is the parameter set of the model.This function aims to maximize the likelihood of accurately predicting responses based on the given instructions.The fine-tuning is performed on eight H100 80G GPUs, over three epochs with a learning rate of 1e−5, a weight decay of 1e−5, a warmup ratio of 0.01, and Low-Rank Adaptation (LoRA) for parameter-effective tuning78。
Output parsing.For extractive and classification tasks, we extracted the targeted predictions (e.g., classification types or multiple-choice options) from the raw outputs of LLMs with a combination of manual and automatic processing.We manually reviewed the processed outputs.Manual review showed that LLMs provided answers in inconsistent formats in some cases.For example, when presenting multiple-choice option C, the raw output examples included variations such as: “Based on the information provided, the most likely … is C. The thyroid gland is a common site for metastasis, and …â€, “Great!Let’s go through the options.A. … B. …Therefore, the most likely diagnosis is C.â€, and “I’m happy to help!Based on the patient’s symptoms and examination findings, … Therefore, option A is incorrect.…, so option D is incorrect.The correct answer is option C.†(adapted from real responses with unnecessary details omitted).In such cases, automatic processing might overlook the answer, potentially lowering LLM accuracy.Thus, we manually extracted outputs in these instances to ensure fair credit.Additionally, we qualitatively evaluated the prevalence of such cases (providing responses in inconsistent formats), which will be introduced below.
评估
Quantitative evaluations.We summarized the evaluation metrics in Table 5under zero-shot, static few-shot, dynamic K-nearest few-shot, and fine-tuning settings.The metrics are applicable to the entire testing sets of 12 datasets.We further conducted bootstrapping using a subsample size of 30 and repeated 100 times at a 95% confidence interval to report performance variance and performed a two-tailed Wilcoxon rank-sum test using SciPy79。Further details are provided in Supplementary Information S2Quantitative evaluation results (S2.1. Result reporting).
Qualitative evaluations on inconsistency, missing information, and hallucinations.For the tasks where the gold standard is fixed, e.g., a classification type or multiple-choice option, we conducted qualitative evaluations on collectively hundreds of thousands of raw outputs of the LLMs (the raw outputs from three LLMs under zero- and one-shot conditions across three benchmarks) to categorize errors beyond inaccurate predictions.Specifically, we examined (1) inconsistent responses, where the responses are in different formats, (2) missingness, where the responses are missing, and (3) hallucinations, where LLMs fail to address the prompt and may contain repetitions and misinformation in the output36。We evaluated and reported the results in selected datasets: ChemProt, HoC, and MedQA.
Qualitative evaluations on accuracy, completeness, and readability.For the tasks with free-text gold standards, such as summaries, we conducted qualitative evaluations on the quality of generated text.Specifically, one senior resident and one junior resident evaluated four models: the fine-tuned BART model reported in the SOTA approach, GPT-3.5 zero-shot, GPT-4 zero-shot, and LLaMA 2 13B zero-shot on 50 random samples from the PubMed Text Summarization benchmark.Each annotator was provided with 600 annotations.To mitigate potential bias, the model outputs were all lowercased, their orders were randomly shuffled, and the annotators were unaware of the models being evaluated.They assessed three dimensions on a scale of 1—5: (1) accuracy, does the generated text contain correct information from the original input, (2) completeness, does the generated text capture the key information from the original input, and (3) readability, is the generated text easy to read.The detailed evaluation guideline is provided in Supplementary Information S3Qualitative evaluation on the PubMed Text Summarization Benchmark.
Cost analysis
We further conducted a cost analysis to quantify the trade-off between cost and accuracy when using GPT models.The cost of GPT models is determined by the number of input and output tokens.We tracked the tokens in the input prompts and output completions using the official model tokenizers provided by OpenAI (https://cookbook.openai.com/examples/how_to_count_tokens_with_tiktoken) and used the pricing table (https://azure.microsoft.com/en-us/pricing/details/cognitive-services/openai-service/) to compute the overall cost.
报告摘要
Further information on research design is available in the Nature Portfolio Reporting Summarylinked to this article.
数据可用性
All data supporting the findings of this study, including source data, are available in the article and Supplementary Information, and can be accessed publicly viahttps://doi.org/10.5281/zenodo.1402550037。Additional data or requests for data can also be obtained from the corresponding authors upon request. 源数据are provided with this paper.
参考
Sayers, E. W. et al.Database resources of the National Center for Biotechnology Information in 2023.核酸res。 51, D29–D38 (2023).
CAS一个 PubMed一个 数学一个 Google Scholar一个
Chen, Q. et al.LitCovid in 2022: an information resource for the COVID-19 literature.核酸res。 51, D1512–D1518 (2023).
PubMed一个 Google Scholar一个
Leaman, R. et al.Comprehensively identifying long COVID articles with human-in-the-loop machine learning.模式 4, 100659 (2023).
Chen, Q. et al.BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale.PLoS Comput.生物。 16, e1007617 (2020).
CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Blake, C. Beyond genes, proteins, and abstracts: Identifying scientific claims from full-text biomedical articles.J. Biomed。通知。 43, 173–189 (2010).
CAS一个 PubMed一个 数学一个 Google Scholar一个
Su, Y. et al.Deep learning joint models for extracting entities and relations in biomedical: a survey and comparison.简短的。Bioinforma。 23, bbac342 (2022).
Zhang, Y., Chen, Q., Yang, Z., Lin, H., & Lu, Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH.科学。数据。 6, 1–9 (2019).
Chen, Q., Peng, Y. & Lu, Z. BioSentVec: creating sentence embeddings for biomedical texts.In2019 IEEE International Conference on Healthcare Informatics (ICHI)1–5 (IEEE, 2019).
Lee,J。等。BioBERT: a pre-trained biomedical language representation model for biomedical text mining.生物信息学 36, 1234–1240 (2020).
CAS一个 PubMed一个 数学一个 Google Scholar一个
Peng, Y., Yan, S., & Lu, Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets.在Proc。18th BioNLP Workshop and Shared Task, 58–65 (Association for Computational Linguistics, Florence, Italy, 2019).
Fang, L., Chen, Q., Wei, C.-H., Lu, Z. & Wang, K. Bioformer: an efficient transformer language model for biomedical text mining, arXiv preprint arXiv:2302.01588 (2023).
Luo,R。等。BioGPT: generative pre-trained transformer for biomedical text generation and mining.简短的。Bioinforma。 23, bbac409 (2022).
Venigalla, A., Frankle, J., & Carbin, M. Biomedlm: a domain-specific large language model for biomedical text, MosaicML。Accessed: Dec, 23 (2022).Yuan, H. et al.
BioBART: Pretraining and evaluation of a biomedical generative language model.在Proc。21st Workshop on Biomedical Language Processing, 97–109 (2022).
Phan, L.N.等。Scifive: a text-to-text transformer model for biomedical literature, arXiv preprint arXiv:2106.03598 (2021).
Gu, Y. et al.Domain-specific language model pretraining for biomedical natural language processing.ACM Trans。计算。Healthc。健康,,,,3, 1–23 (2021).
Allot, A. et al.LitSense: making sense of biomedical literature at sentence level.核酸res。 47, W594–W599 (2019).
CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zhao, W. X. et al.A survey of large language models, arXiv preprint arXiv:2303.18223 (2023).
Ouyang,L。等。培训语言模型遵循人类反馈的说明。ADV。神经信息。过程。系统。 35, 27730–27744 (2022).
数学一个 Google Scholar一个
Chen,X。等。How Robust is GPT-3.5 to Predecessors?A Comprehensive Study on Language Understanding Tasks, arXiv preprint arXiv:2303.00293 (2023).
OpenAI, GPT-4 Technical Report, ArXiv, abs/2303.08774, (2023).
Touvron,H。等。Llama 2: Open foundation and fine-tuned chat models, arXiv preprint arXiv:2307.09288 (2023).
Jiang, A. Q. et al.Mixtral of experts arXiv preprint arXiv:2401.04088, 2024.
Lee, P, Goldberg, C. & Kohane, I. The AI revolution in medicine: GPT-4 and beyond (Pearson, 2023).
Wong, C. et al.Scaling clinical trial matching using large language models: A case study in oncology.在Machine Learning for Healthcare Conference846–862 (PMLR, 2023).
Liu,Q。等。Exploring the Boundaries of GPT-4 in Radiology.在Proc。of the 2023 Conference on Empirical Methods in Natural Language Processing14414–14445 (2023).
Nori, H. et al.Can generalist foundation models outcompete special-purpose tuning?Case study in medicine, arXiv preprint arXiv:2311.16452 (2023).
Tian,S。等。Opportunities and challenges for ChatGPT and large language models in biomedicine and health.简短的。Bioinforma。 25, bbad493 (2024).
He, K. et al.A survey of large language models for healthcare: from data, technology, and applications to accountability and ethics, arXiv preprint arXiv:2310.05694 (2023).
Omiye, J. A., Gui, H., Rezaei, S. J., Zou, J. & Daneshjou, R. Large language models in medicine: the potentials and pitfalls: a narrative review.安。实习生。医学 177, 210–220 (2024).
PubMed一个 Google Scholar一个
SoÄŸancıoÄŸlu, G., Öztürk, H. & Özgür, A. BIOSSES: a semantic sentence similarity estimation system for the biomedical domain.生物信息学 33, i49–i58 (2017).
Tinn, R. et al.Fine-tuning large neural language models for biomedical natural language processing.Patterns. 4, 100729 (2023).
Chen, Q., Rankine, A., Peng, Y., Aghaarabi, E. & Lu, Z. Benchmarking effectiveness and efficiency of deep learning models for semantic textual similarity in the clinical domain: validation study.Jmir Med。通知。 9, e27386 (2021).
Wu, C. et al.PMC-LLaMA: toward building open-source language models for medicine,J. Am。医学通知。Associat。ocae045 (2024).
Fleming, S. L. et al.MedAlign: A clinician-generated dataset for instruction following with electronic medical records.在Proc。AAAI人工智能会议卷。38 22021–22030 (2023).
Zhang,Y。等。Siren’s song in the AI ocean: a survey on hallucination in large language models.arXiv preprint arXiv:2309.01219 (2023).
Chen, Q. et al.A systematic evaluation of large language models for biomedical natural language processing: benchmarks, baselines, and recommendations.https://doi.org/10.5281/zenodo.14025500(2024)。
Nori, H., King, N., McKinney, S. M., Carignan, D. & Horvitz, E. Capabilities of gpt-4 on medical challenge problems.arXiv preprint arXiv:2303.13375 (2023).
Labrak, Y., Rouvier, M. & Dufour, R. A zero-shot and few-shot study of instruction-finetuned large language models applied to clinical and biomedical tasks.在Proc。2024 Joint International Conference on Computational Linguistics, Language Resources and Evaluation (LREC-COLING 2024)2049–2066 (ELRA and ICCL, 2024).
Jin, H. et al.Llm maybe longlm: Self-extend llm context window without tuning.在Proc。of Machine Learning Research,,,,23522099–22114 (2024).
Ding, Y. et al.LongRoPE: Extending LLM Context Window Beyond 2 Million Tokens, arXiv preprint arXiv:2402.13753 (2024).
Xie, Q., Huang, J., Saha, T. & Ananiadou, S. Gretel: Graph contrastive topic enhanced language model for long document extractive summarization.在Proc。29th International Conference on Computational Linguistics, 6259–6269 (International Committee on Computational Linguistics, 2022).
Jimenez Gutierrez, B. et al.Thinking about GPT-3 in-context learning for biomedical IE?想一想。在Findings of the Association for Computational Linguistics: EMNLP 2022, 4497–4512 (Association for Computational Linguistics, 2022).
Rehana, H. et al.Evaluation of GPT and BERT-based models on identifying protein-protein interactions in biomedical text, arXiv preprint arXiv:2303.17728 (2023).
Jin, Q. et al.Biomedical question answering: a survey of approaches and challenges.ACM计算。Surv.(CSUR) 55, 1–36 (2022).
数学一个 Google Scholar一个
Singhal, K. et al.Large language models encode clinical knowledge,自然 620, 1–9 (2023).
Chang,Y。等。A survey on evaluation of large language models,ACM Trans。Intell。系统。技术。(2023)。
Minaee, S. et al.Large language models: A survey, arXiv preprint arXiv:2402.06196 (2024).
Chen,Z。等。Meditron-70b: Scaling medical pretraining for large language models, arXiv preprint arXiv:2311.16079 (2023).
DeYoung, J., Beltagy, I., van Zuylen, M., Kuehl, B. & Wang, L. L. Ms2: Multi-document summarization of medical studies.在Proc。2021 Conference on Empirical Methods in Natural Language Processing, 7494–7513 (2021).
Wornow, M. et al.The shaky foundations of large language models and foundation models for electronic health records.npj Digit.医学 6, 135 (2023).
Manyika, J. An overview of Bard: an early experiment with generative AI.https://ai.google/static/documents/google-about-bard.pdf(2023)。
Jiang, A. Q. et al.Mistral 7B, arXiv preprint arXiv:2310.06825, (2023).
Neelakantan, A. et al.Text and code embeddings by contrastive pre-training, arXiv preprint arXiv:2201.10005 (2022).
Krallinger, M. et al.Overview of the BioCreative VI chemical-protein interaction Track.在Proc。of the sixth BioCreative challenge evaluation workshop卷。1, 141–146 (2017).
Chen, Q. et al.Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations,数据库 2022, baac069 (2022).
Islamaj DoÄŸan, R. et al.Overview of the BioCreative VI Precision Medicine Track: mining protein interactions and mutations for precision medicine,数据库 2019, bay147 (2019).
International Society for Biocuration, Biocuration: Distilling data into knowledge,Plos Biol.,,,,16, e2002846 (2018).
Li, J. et al.BioCreative V CDR task corpus: a resource for chemical disease relation extraction,数据库, 2016 (2016).
DoÄŸan, R. I., Leaman, R. & Lu, Z. NCBI disease corpus: a resource for disease name recognition and concept normalization.J. Biomed。通知。 47, 1–10 (2014).
PubMed一个 PubMed Central一个 数学一个 Google Scholar一个
Li, X., Rousseau, J. F., Ding, Y., Song, M. & Lu, W. Understanding drug repurposing from the perspective of biomedical entities and their evolution: Bibliographic research using aspirin.Jmir Med。通知。 8, e16739 (2020).
Segura-Bedmar, I., MartÃnez, P. & Herrero-Zazo, M. Semeval-2013 task 9: extraction of drug-drug interactions from biomedical texts (ddiextraction 2013).在Second Joint Conference on Lexical and Computational Semantics (* SEM), Volume 2: Proc.Seventh International Workshop on Semantic Evaluation (SemEval 2013)341–350 (Association for Computational Linguistics, 2013).
Du, J. et al.ML-Net: multi-label classification of biomedical texts with deep neural networks.J. Am。医学通知。联合。 26, 1279–1285 (2019).
PubMed一个 PubMed Central一个 数学一个 Google Scholar一个
Baker, S. et al.Automatic semantic classification of scientific literature according to the hallmarks of cancer.生物信息学 32, 432–440 (2016).
CAS一个 PubMed一个 数学一个 Google Scholar一个
Kaddari, Z., Mellah, Y., Berrich, J., Bouchentouf, T. & Belkasmi, M. G. Biomedical question answering: A survey of methods and datasets.在2020 Fourth International Conference On Intelligent Computing in Data Sciences (ICDS)1–8 (IEEE, 2020).
Jin, D. et al.What disease does this patient have?A large-scale open domain question answering dataset from medical exams.应用。科学。 11, 6421 (2021).
CAS一个 数学一个 Google Scholar一个
Jin, Q., Dhingra, B., Liu, Z., Cohen, W. & Lu, X. Pubmedqa: A dataset for biomedical research question answering.在Proc。2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, 2567–2577 (EMNLP-IJCNLP, 2019).
Cohan, A. et al.A discourse-aware attention model for abstractive summarization of long documents.在Proc。2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Languag Technologies卷。2, 615–621 (2018).
Devaraj, A., Wallace, B. C., Marshall, I. J. & Li, J. J. Paragraph-level simplification of medical texts.在Proc。2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies4972–4984 (Association for Computational Linguistics, 2021).
Luo, Z., Xie, Q., & Ananiadou, S. Readability controllable biomedical document summarization.在Findings of the Association for Computational Linguistics: EMNLP, 4667–4680 (2022).
Goldsack, T. et al.Overview of the biolaysumm 2024 shared task on lay summarization of biomedical research articles.在Proc。23rd Workshop on Biomedical Natural Language Processing122–131 (2024).
Lewis, M. et al.Bart: Denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension.在Proc。58th Annual Meeting of the Association for Computational Linguistics, 7871–7880 (2020).
Liu,P。等。Pre-train, prompt, and predict: A systematic survey of prompting methods in natural language processing.ACM计算。Surv. 55, 1–35 (2023).
数学一个 Google Scholar一个
Hu,Y。等。Improving large language models for clinical named entity recognition via prompt engineering,J. Am。医学通知。联合。 31, ocad259 (2024).
Wang,L。等。Investigating the impact of prompt engineering on the performance of large language models for standardizing obstetric diagnosis text: comparative study.JMIR Format Res. 8, e53216 (2024).
Agrawal, M., Hegselmann, S., Lang, H., Kim, Y. & Sontag, D. Large language models are few-shot clinical information extractors.在Proc。2022 Conference on Empirical Methods in Natural Language Processing, 1998–2022 (2022).
Keloth, V. K. et al.Advancing entity recognition in biomedicine via instruction tuning of large language models.生物信息学 40, btae163 (2024).
CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Hu, E. J. et al.Lora: Low-rank adaptation of large language models, arXiv preprint arXiv:2106.09685 (2021).
Virtanen, P. et al.SciPy 1.0: fundamental algorithms for scientific computing in Python.纳特。方法 17, 261–272 (2020).
CAS一个 PubMed一个 PubMed Central一个 数学一个 Google Scholar一个
Lehman, E. et al.Do we still need clinical language models?In Conference on health, inference, and learning, 578–597 (PMLR, 2023).
Chen,S。等。Evaluating the ChatGPT family of models for biomedical reasoning and classification.J. Am。医学通知。联合。 31, ocad256 (2024).
Chen, Q. et al.A comprehensive benchmark study on biomedical text generation and mining with ChatGPT,Biorxiv, pp. 2023.04.19.537463 (2023).
Zhang, S., Cheng, H., Gao, J. & Poon H. Optimizing bi-encoder for named entity recognition via contrastive learning.在Proc。11th International Conference on Learning Representations, (ICLR, 2023).
He, J. et al.Chemical-protein relation extraction with pre-trained prompt tuning.Proc IEEE Int.conf。Healthc。通知。2022, 608–609 (2022).Mingliang, D., Jijun, T. & Fei, G. Document-level DDI relation extraction with document-entity embedding.
pp. 392–397.
Chen, Q., Du, J., Allot, A. & Lu, Z. LitMC-BERT: transformer-based multi-label classification of biomedical literature with an application on COVID-19 literature curation,IEEE/ACM Trans。计算。生物。Bioinform。19, 2584–2595 (2022).Yasunaga, M. et al.
Deep bidirectional language-knowledge graph pretraining.ADV。神经信息。过程。系统。 35, 37309–37323 (2022).
Flores, L. J. Y., Huang, H., Shi, K., Chheang, S. & Cohan, A. Medical text simplification: optimizing for readability with unlikelihood training and reranked beam search decoding.在Findings of the Association for Computational Linguistics: EMNLP, 4859–4873 (2023).
Wei, C.-H.等。Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task.数据库 2016, baw032 (2016).
He, J. et al.Prompt tuning in biomedical relation extraction,J. Healthcare Inform.res。 8, 1–19 (2024).
Guo, Z., Wang, P., Wang, Y. & Yu, S. Improving small language models on PubMedQA via Generative Data Augmentation,arxiv,,,,12(2023)。
Koh, H. Y., Ju, J., Liu, M. & Pan, S. An empirical survey on long document summarization: Datasets, models, and metrics.ACM计算。Surv. 55, 1–35 (2022).
数学一个 Google Scholar一个
Bishop, J. A., Xie, Q. & Ananiadou, S. LongDocFACTScore: Evaluating the factuality of long document abstractive summarisation.在Proc。of the 2024 Joint International Conference on Computational Linguistics, Language Resources and Evaluation (LREC-COLING 2024)10777–10789 (2024).
Wang, L. L, DeYoung, J. & Wallace, B. Overview of MSLR2022: A shared task on multidocument summarization for literature reviews.在Proc。Third Workshop on Scholarly Document Processing175–180 (Association for Computational Linguistics, 2022).
Ondov, B., Attal, K. & Demner-Fushman, D. A survey of automated methods for biomedical text simplification.J. Am。医学通知。联合。 29, 1976–1988 (2022).
PubMed一个 PubMed Central一个 数学一个 Google Scholar一个
Stricker, J., Chasiotis, A., Kerwer, M. & Günther, A. Scientific abstracts and plain language summaries in psychology: A comparison based on readability indices.PLOS一个 15, e0231160 (2020).
CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
致谢
This study is supported by the following National Institutes of Health grants: 1R01LM014604 (Q.C., R.A.A., and H.X), 4R00LM014024 (Q.C.), R01AG078154 (R.Z., and H.X), 1R01AG066749 (W.J.Z), W81XWH-22-1-0164 (W.J.Z), and the Intramural Research Program of the National Library of Medicine (Q.C., Q.J., P.L., Z.W., and Z.L).
资金
Open access funding provided by the National Institutes of Health.
道德声明
竞争利益
Dr. Jingcheng Du and Dr. Hua Xu have research-related financial interests at Melax Technologies Inc. The remaining authors declare no competing interests.
同行评审
同行评审信息
自然通讯thanks the anonymous reviewers for their contribution to the peer review of this work.提供同行评审文件。
附加信息
Publisher’s note关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。
补充信息
权利和权限
开放访问本文允许以任何媒介或格式的使用,共享,适应,分发和复制允许使用,分享,适应,分发和复制的国际许可,只要您适当地归功于原始作者和来源,就可以提供与创意共享许可证的链接,并指出是否进行了更改。The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.要查看此许可证的副本,请访问http://creativecommons.org/licenses/4.0/。重印和权限
引用本文
Chen, Q., Hu, Y., Peng, X.
等。Benchmarking large language models for biomedical natural language processing applications and recommendations.纳特社区16 , 3280 (2025).https://doi.org/10.1038/s41467-025-56989-2
已收到:
公认:
出版:
doi:https://doi.org/10.1038/s41467-025-56989-2