癌症磁共振成像中人工智能的批判性评估
作者:Yankeelov, Thomas E.
介绍
我们试图在癌症的磁共振成像(MRI)的背景下对人工智能(AI)的机会和局限性进行批判性评估,并为读者提供有关更多信息的指导。确实,关于MRI中AI的各个方面的出色出版物有许多出色的出版物(例如,参见参考文献1,,,,2,,,,3,,,,4)。但是,考虑到有关该主题的出版物的爆炸,将炒作与实际成功区分开并确定该领域的最新技术可能是一项挑战。特别是,很难确定当前可用的内容以及在癌症MRI中应用AI的实际限制。在这项贡献中,我们试图谨慎地提供对临床肿瘤学内MRI的效用和局限性的更平衡的呈现。为了解决这个问题,我们首先描述了通常在医学成像中经常使用的AI的关键组成部分,尤其是MRI。在第3节中,MRI采集和重建作为回归任务,涉及从原始扫描仪测量值估算图像的体素值。在第4节中,我们讨论了图像分割的分类任务,该任务需要将每个体素分为(例如)肿瘤或健康组织,以及登记,这需要在空间上将图像与公共空间相提并论。第5和6节涉及分别从MRI数据进行诊断和预后的分类任务,这些任务需要考虑到全球特征(例如,肿瘤的位置和大小)以进行预测。
密钥AI概念用于医学成像
什么是人工智能?
人工智能(AI)是指计算机系统的理论和开发,这些系统通常可以执行通常认为需要人类智能的任务,包括语言,视觉感知和推理5。我们专注于称为机器学习(ML)的AI分支,该分支是指培训有关相关数据的统计模型以执行任务。深度学习是ML的子分支,涉及神经网络的发展(NN),这是一种特殊的模型,在医学成像中证明了实用性6,,,,7,,,,8。
常见的ML医学成像技术
开发ML模型时的关键设计考虑因素是可用于培训的数据的类型,可以将其分为监督和无监督的设置。监督数据集包含匹配的{示例,标签}对;例如,注释为包含或不包含肿瘤的MRI图像。在这种情况下,该模型学会了预测每个样本的标签。但是,作为域专家(例如,放射科医生)必须标记每个单独的数据点,注释可能是耗时且昂贵的9。相反,无监督的数据集仅包含没有任何标签的样品。例如,给定一组MR训练图像以低空间分辨率获得,可以训练模型以学习增加分辨率(称为超分辨率10,,,,11)无需访问任何高分辨率示例。训练后,该模型可用于提高以较低分辨率获取的新图像的质量,从而减少了获取高分辨率数据的需求。尽管无监督的设置减轻了注释数据的负担,但生成有用标签的能力与收集与预期应用相关的高质量数据的能力紧密相关。例如,如果仅膝盖图像可用于训练超分辨率模型,则该模型在增强大脑图像时可能会表现不佳12。
读者可能会发现参考表很有帮助1当他们考虑其余的论文时。
基于AI的图像获取和MRI数据的重建
图像采集和重建在MRI中密不可分。图像采集包括信号和空间编码通过用户控制的射频和梯度波形组成。通过修改采集过程的不同组件(脉冲时间延迟,信号频率,信号强度,信号相等),更改了采集速度以及给定图像对特定组织特性的敏感性。所得图像对比可以通过:
在哪里x cn是(矢量化的)图像(包含n基于时空函数生成的体素)f两个采集控制的信号编码参数我和生物物理组织参数,问13。获得的测量(称为K空间)通过线性操作员与图像有关
$$ y = {a} _ {{\ rm {\ phi}}} x+\ eta $$
(2)
在哪里一个 - c曼表示(可能是多层)采样的傅立叶变换,我· cm是添加剂复合物值高斯噪声,并且m是通过空间编码参数确定的位置的获得的测量数量 -。测得的原始信号(即
y)未准备好可视化,因为它们代表图像的傅立叶组件。因此,优化算法用于重建图像。重建算法可以用函数表示g((y)将线性测量过程反转。((2)估计图像x,或反转等式中的非线性测量过程。((1)为了估计组织参数,问。这些任务通常是具有挑战性的,因为数据被添加剂噪声损坏并经常进行(经常采样)(m<<n)减少扫描时间,从而导致不良反相问题。图像采集和重建中的常见机器学习技术
培训机器学习模型以协助采集和重建程序,通常被构成优化问题,我们在其中训练参数化功能
\({f} _ {w}(\ cdot)\),通常以深层神经网络的形式完成特定任务。在监督的情况下,给定输入K空间的训练图像(或扫描参数或组织参数)相对于训练图像(或扫描参数或组织参数)进行了优化。这通常采用最小化某些损失功能的形式\(d \ left(\ cdot,\ cdot \ right)\)\)通过以下方式通过大规模优化求解器:
$ {w}^{*} = {argmi} {n} _ {w} \ mathop {\ sum} \ limits_ {i = 1}^{n} d \ left({f} _ {w} _ {w} _ {w}
(3)
在哪里x我和y我代表我训练样本和n是训练集的大小。在无监督的学习中,解决了相同的任务,但最小化仅取决于输入y我。图像获取信号编码参数的选择我
和空间编码参数
-可以影响所得图像的对比度(等式(等式)((1)和收购时间(等式((等式)(2)。由于对病理的图像敏感性受到习得参数的影响,因此优化图像采集管道以增加对相关组织对比的敏感性是有意义的。这意味着找到最佳的射频翻转角,相,时机等,以进行所需的对比度。监督学习已被用来寻找这些采集策略来优化对比度灵敏度14,,,,15以及优化K空间测量16,,,,17,,,,18,,,,19。这里w在等式中。((3)由对比的任何一个或两个都取代(我)和k空间位置( -)。例如,k空间轨迹优化可用于通过减少获得的数量来加速成像k - 给定目标空间分辨率的空间测量。在这种情况下,损失功能将采用表格
$$ {\ phi}^{*} = {argmi} {n} _ {\ phi} \ mathop {\ sum} \ limits_ {i = 1}^{n} d {n} d \ lest({f})\ right),{x} _ {i} \ right)$$
(4)
在哪里y我( - )代表K空间中特定的采样轨迹(例如,特定相位编码线)我训练样本,以及\({f} _ {w} \ left({y} _ {i} \ left(\ phi \ right)\ right)\ right)\)\)代表一种重建算法,该算法采用了子采样并输出图像。当重建网络和采样轨迹都是可区分的时,这很容易实现。但是,对于笛卡尔抽样,这个问题本质上是组合的。因此,必须采用贪婪的方法或平滑近似16,,,,17,,,,18,,,,19)。可以使用类似的方法来更新影响图像对比的其他扫描参数15。
等式之后。((4解决),可以将优化的采集方案表示为新的测量模型一个 -并且可以在扫描仪上实现。当使用此序列进行获取时,鉴于获得的测量和测量模型,可以解决图像重建问题(即等式。2)。
图像重建
经典图像重建依赖于手工制作的先验l1 - 大波正则化20。最近,深度神经网络为图像重建提供了有希望的技术。基于AI的技术可以分为几类:端到端监督,端到端无监督和生成的建模。在端到端监督设置中,功能fw在等式中。((3)经过成对的子采样训练k - 从完全采样的数据获得的空间数据和图像((y我,,,,x我), 和d(!½¥,§¥)可以代表任何有效的距离度量(例如,平方误差)。端到端的无监督方法还训练NN,除了仅访问亚采样测量,y我,没有随附的参考图像,x我。这在只能收集大量的亚采样数据(例如,在动态成像中)的情况下很重要。用于监督和无监督的重建的端到端NN可以采用多种形式,但是展开的优化网络在经典优化步骤(例如,共轭梯度下降)之间交替出21,,,,22。尽管端到端的方法具有强大的功能,但性能会随着在训练时进行测量与测试时的测量方式的变化,这被称为测试时间分配变化。最近,已经证明将生成模型用于逆问题可改善对采集方案的变化的鲁棒性。
这些方法训练贝叶斯的先验,就完全采样的图像的分布,p((x),因此对分布的可能性变化不可知,p((y|x),可以在扫描和成像协议(即信号和空间编码参数)中进行更改。也许最流行的方法是通过基于分数或扩散概率模型来包括逆问题中的生成先验23,,,,24。
尽管有监督和无监督训练之间有区别,但必须注意的是,AI模型是根据原始传感器(即原始K-空间测量值)培训的,这些数据通常是通过多线圈阵列获取的,并且通常是对的。即使数据完全采样,它们也很嘈杂,必须首先将其重建为图像。因此,没有真正的基础图像的概念。已经提出了解释采集过程的新培训方法,既是针对经过培训的端到端方法,这些方法经过训练以进行获得的测量和输出重建图像,以及生成模型(其中AI模型在迭代重建之前用作统计图像)。
也可以与重建方法一起共同训练采集参数,这可能由深神经网络组成14,,,,15,,,,16,,,,17。例如,Aggarwal等人,将优化问题提出为
$$ {\ phi}^{*} {w}^{*} = {argmi} {n} {n} _ {\ phi,w} \ mathop {\ sum {\ sum {\ sum} \ limits_ {i = 1}^{n} d \ left({f} _ {w} \ left({y} _ {i} _ {i}(\ phi)\ right),{x} _ {i} _ {i}
(5)
换句话说,他们共同解决了由 -以及由fw。
当前基于AI的图像获取和重建的临床需求
AI表明,使用较少的测量值比传统的重建技术保持了高图像质量的能力,甚至可以产生用于特定下游临床任务的诊断图像25。例如,与传统的重建方法相比26。图像重建的AI已进入了几家扫描仪制造商的产品。最近有两个例子是盖的空气TM重建协议和西门的深刻决心TM产品线。但是,重要的是要强调,其中许多产品用于专业案例(例如,特定的解剖学/对比度),目前尚未针对许多特定用例部署。跨成像协议概括的能力对于广泛采用至关重要。
许多更长的扫描课程的另一个问题是患者运动,尤其是针对小儿种群。通常,这包括开发可以处理动态成像场景的强大重建方法(例如,对比增强成像27)从单个成像量中收集所需的测量通常是不可行的。由于许多形式的病理学仅表现出细微的对比变化,因此还需要获得能够解决组织特性的细微变化的获取/重建方法,从而减少了含有Gadolinium的外源性对比剂的使用和剂量28。最后,由于许多新方法通常仅基于图像质量指标进行训练和衡量性能,因此必须更好地量化图像质量和下游诊断指标(例如肿瘤分类)之间的联系,并且在设计新的基于AI的技术时必须更好地理解和优化。
在临床环境中实践部署的障碍
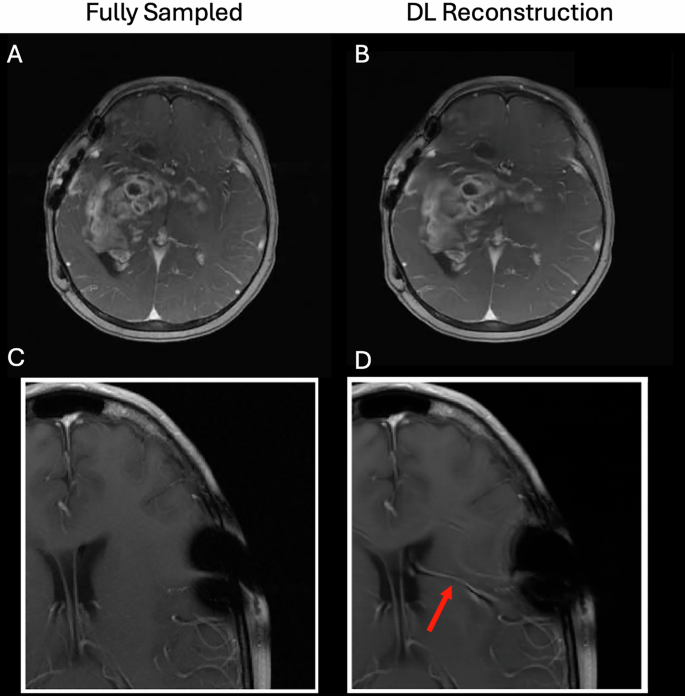
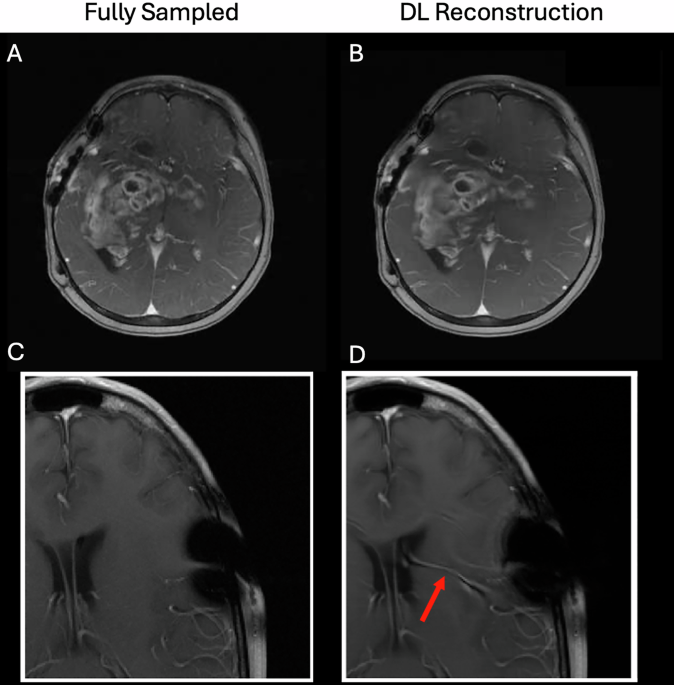
尽管AI对经典的重建和获取技术表现出明显的改进17,,,,21,,,,22,,,,23,在提出临床采用之前,仍然需要解决许多问题。例如,由于供应商,现场强度,现场不均匀性和患者运动的差异,AI技术对于如何收集数据的变化需要鲁棒。必须对硬件和成像协议中每个变化的代表性数据进行培训;如果扫描协议更改(或者扫描参数更改以适应特定患者,例如较大的视野,不同的分辨率,不同的回声时间,较大的脂肪饱和度频段等),那么该新协议可以超出训练集中所包含的分布,并且随后的性能将在Jalal等人中脱落。23。这可以迅速转变为模型汤。1。还必须通过不确定性定量来提高可解释性。为了证明这些要点,图。1显示两个示例深度学习重建(从2020年FastMRI重建挑战中复制29)回顾性扫描完全采样以模拟更快的扫描。在第一种情况下,DL重建会导致忠实的诊断图像;然而,在第二种情况下,DL重建错误使血管虚假幻觉,这可能是由于手术钉书钉引起的看不见的人造物所致。这证明了在临床环境中部署这些模型的巨大挑战,并且可能存在临床上的困惑问题30。
换句话说,生成模型经过训练,以学习独立于产生这些图像的物理成像系统参数的MR图像的分布。这意味着生成模型可用于从其他成像方案中获取的MR数据中重建图像,只要图像遵循模型的学习分布,并且已知成像参数。学习生成模型的最佳抽样模式也可能会有所帮助,因为这将使获取更加针对特定的成像协议,同时仍然受益于基于生成模型的重建所提供的鲁棒性。
注册和细分MRI数据通过人工智能
分段和注册中的常见AI技术
肿瘤和组织分割
肿瘤和组织分割通常用于手术计划,放疗设计和评估治疗反应31,,,,32。目前,手动或半自动化的分段通常会遭受资源密集型和不可接受的观察者间变异性的损失31,,,,33。尽管AI启用了自动细分方法可能会减少这两种问题,但它们需要具有专家定义的细分的广泛培训集,并且(由于其黑色盒子的性质)可能会在没有警告的情况下失败31,,,,34。鉴于这些限制,基于AI的方法需要人类的监督来审查,并在必要时进行编辑31,,,,35。许多AI自动化方法基于基于体素的特征,通常采用卷积神经网络(CNN;例如U-NET)来鉴定健康组织(例如,用于放射疗法计划的器官风险),癌症和肿瘤内子区域(例如,肿瘤内的子区域(例如,肿瘤,湿疹,湿疹,增强病情)32。Pal等。引入了一种结合模糊c均值聚类和随机森林算法的方法,并显示出99%的精度用于分割脊柱肿瘤36。Kundal等。评估了四种基于CNN的脑肿瘤分割方法。(Captk,2Dvnet,Ensememunets和Resnet50)Ensememunets的骰子得分为0.93和0.85,Hausdorff距离分别为18和17.5,用于测试和验证37。尽管当前的方法并不能消除对人类精致的需求,但它们通常可以提供有风险的器官的可接受分割,当需要更高的精度和准确性时,可以手动改进38。
图像注册
经典图像注册方法包括估计最大化一组图像与目标图像之间相似性的最佳转换。存在转换(刚性或可变形)和成本函数(总平方差,归一化的互相关或互信息)的多种组合。Osman等。引入了可变形CNN的3D MRI扫描的胶质瘤患者的注册39这表现优于VoxelMorph方法,平均骰子得分为0.975,相似性指数为0.908,而0.969和0.893。但是,每种方法都有特定的缺点,例如对工件敏感或捕获局部差异时受到限制40。基于学习的方法是受监督和无监督的,试图克服这些困难。有监督的学习模型,例如Birnet41和Deepflash42使用配对图像(即注册和未注册的图像)进行培训。即使此类方法可以实现准确的注册性能,但很难找到必要的培训数据(包括注册图像和相关的转换)。因此,无监督的深度学习方法是首选43,,,,44传统上,使用基于梯度的优化器(如随机梯度下降)来优化相似性指标和模型架构。
当前基于AI的图像细分和注册的临床需求
我们为基于DL的脑癌MRI登记分配了适度的准备状态。特别是,在肿瘤边界处的局部注册体素很具有挑战性。埃斯蒂安等人。45引入了一种联合3D-CNN方法,用于脑登记和肿瘤分割。使用Brats 2018数据集对该模型进行了培训46然后从同一数据集对200多对测试。通过计算两个比率之间的平均距离来评估注册方法:(1)原始到变形的肿瘤面膜面积,以及(2)成对图像的脑体积之间。该方法的表现超过了使用整个肿瘤面膜的先前建立的方法(VoxelMorph软件包的一部分47)采用了一种基于学习的推理算法,该算法最初应用于健康的大脑MRIS48。
在几个领域,基于DL的细分可以满足临床需求。首先,从周围的健康组织中分割肿瘤是放疗计划的核心。实际上,手动细分的时间密集型性质限制了适应性放射疗法的广泛实施49。为了应对这一挑战,已经开发了使用CNN执行半自动化和自动分段的AI工具。这样的工具通常在治疗后的性能降低(即手术,放射疗法和化学疗法)。50使肿瘤的准确纵向分割。这些自动分割工具的临床部署旨在加速自适应放射疗法工作流程中的分割任务。但是,这些算法因其在培训期间没有看到的数据时的可推广性有限而取得了不同的成功,因此限制了其广泛采用。缺乏基于AI的分割算法成功的潜在原因,包括扫描获取的变异性,正常组织和肿瘤的患者特定表现以及治疗临床团队的分割目标。
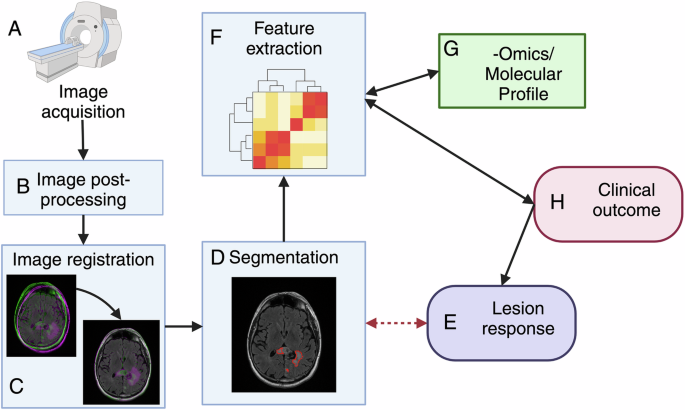
肿瘤分割和注册也是定量描述代表潜在生物学的成像特征的必要步骤(图。2)。MRI分割的不一致可能是由不同读者进行的分割之间的各种因素引起的,以及在同一读者完成的分割中多次分割的评估者变异性多次。不一致的其他来源包括因获得协议,不同分割方法和/或参数设置的算法偏差而引起的输入数据质量和解决方案的变化,解剖结构的时间变化或由于疾病进度效应造成的分割目标的强度对比度51,,,,52,,,,53,,,,54。这些不一致引起了分段区域的体积,形态和对比度的偏见,并影响(例如)放射线特征的准确且可重复的提取55,,,,56,,,,57,,,,58,这限制了放射素学在诊断和预后中临床应用的可靠性和普遍性59,,,,60。因此,开发方法以提高分割的一致性以提高定量成像是至关重要的。
当前注册和细分方法的缺点
当前基于AI的细分和注册方法的根本缺点是可用数据本身培训和验证这些方法的局限性。这些局限性包括在目标设置中缺乏数据,数据质量的变化,成像协议的标准化或成像协议与用于训练先前验证的方法的成像协议的偏差。例如,在脑癌的设置中,虽然历史上有重大努力为细分术前肿瘤,但最近对180篇文章的文献综述只观察到三篇论文,其中包括术后成像31缺乏训练有素的模型或专家细分或精选的数据限制61注册和分割方法转移到治疗可能会改变图像对比度或引入解剖结构的大变形和改变的新应用的可转移性。另一个陷阱是数据漂移的现象,其中成像协议或其他因素的变化可能导致输入数据,而输入数据不在用于训练的分布之外62导致分割和注册的精度降低。
通过MRI数据诊断的AI
常见的AI诊断技术通过MRI
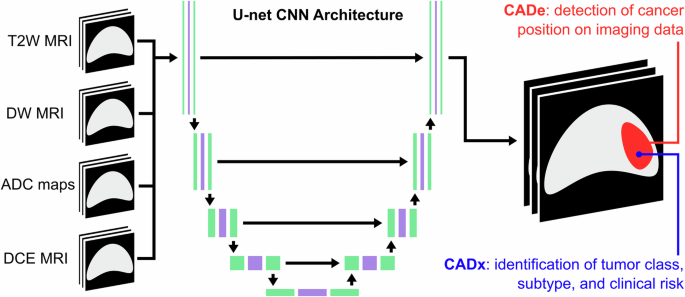
计算机辅助检测系统(CADE)利用AI识别肿瘤在图像中的位置,而计算机辅助诊断(CADX)系统则利用成像特征来定量表征它(见图。3)。这些技术是使用基于放射线学的AI方法构建的32,,,,63,,,,64,,,,65(例如,随机森林,支持向量机)和DL32,,,,63,,,,64,,,,65。特别是,CNN是基于DL的CADE和CADX的最常见架构选择。例如,Chakrabarty等。66培训了一个有CNN,以对比后,t1 - 对六种脑肿瘤类型和健康组织进行鉴别诊断的加权数据,在内部和外部验证中达到了0.95以上的AUC。此外,Saha等人。67使用基于CNN的CADE/CADX系统实现了超过0.86的AUC,用于临床上重要的前列腺癌。t2 - 加权,扩散加权MRI(DW-MRI),明显的扩散系数图(ADC)和区域癌症患病率的解剖学先验。尽管CNN培训需要大量标记的数据集和计算资源,但预先培训的CNN(例如Alexnet,Googlenet)也可以在新数据集中用于癌症检测和诊断任务(即转移学习)32,,,,65,,,,68,,,,69。例如,Antropova等。70在动态对比增强的MRI(DCE-MRI)数据上,与CADX系统诊断出恶性乳腺病变的AUC为0.89,该数据与CADX系统相结合,将放射素与基于CNN的特征相结合,他们使用了预训练的VGG19模型。
当前的临床需求
正在进行的努力来利用AI分析癌症检测和诊断的标准MRI数据,以减少阅读器间的可变性,同时还简化了时间密集型诊断过程32,,,,63,,,,64,,,,65,,,,69。这些方法试图及时可靠地帮助MRI信息,以诊断,监测和治疗。例如,CADE/CADX系统可以帮助确定适当的临床管理策略,以鉴定临床上重要的前列腺癌64,,,,69在多参数MRI数据上。神经肿瘤学的基本挑战是对原发性中枢神经系统肿瘤亚型和脑转移的鉴别诊断,这需要不同的治疗方法32,,,,66。乳腺癌诊断的当前临床需求包括早期发现高风险疾病,改善中等风险和致密乳房种群的筛查,良性病变和恶性病变之间的分化以及特定亚型的鉴定63,,,,65,,,,70,,,,71。这些都是可能通过利用AI方法来帮助的所有任务,尽管其发展面临一些挑战(请参见第5.3节)。此外,最新的努力是针对使用AI更好地告知癌症管理的多模式数据(例如,成像,组织病理学,生物标志物,OMICS)的整合。
Barriers to practical deployment in the clinical setting
In Medicine, as opposed to many other fields of application of AI, understanding why a decision is made (e.g., a diagnosis) is as important as the decision itself.Thus, establishing model interpretability is a fundamental barrier in the clinical translation of AI-based techniques for decision-making in clinical oncology63,,,,64,,,,72,,,,73。Beyond explaining the biophysical causes underlying AI-driven cancer detection and diagnosis, model interpretability is also required to extrapolate and interrogate model outcomes63,,,,64, which would contribute to more informed clinical decisions.The medical AI community has proposed several approaches to address the lack of interpretability in MRI-informed AI models for cancer detection and diagnosis74,,,,75,,,,76, such as class activation methods66,,,,67, knowledge-driven priors67,,,,74, and integration of multimodal multiscale data77,,,,78。Furthermore, recent approaches in the field of scientific machine learning have shown promise in improving the interpretability of AI models79and could therefore be developed for the detection and diagnosis of cancer on MRI data.For example, mechanistic feature engineering can identify biophysically-relevant inputs that characterize tumor biology80,,,,81, physics-informed neural networks (PINNs)82,,,,83include a (bio)physical model in the loss function, and biology-informed neural networks (BINNs) adapt the model architecture according to prior biological knowledge84,,,,85。
Despite the progress in the development of MRI-informed AI methods for cancer detection and diagnosis, these technologies need to address potentially critical pitfalls in their performance in tumor-specific scenarios.For example, the detection of malignant breast lesions using MRI-informed AI models can be affected by complex tumor geometries (e.g., non-mass tumors), the architectural and radiological features of surrounding healthy tissue (e.g., tissue density, background parenchymal enhancement), as well as signal distortion and movement of the tumor during DCE acquisition65,,,,71,,,,86,,,,87。Additionally, the performance of CADe/CADx methods for prostate cancer can be affected by several well-established MRI confounders64,,,,67,,,,88,,,,89,,,,90, such as prostatitis and benign prostatic hyperplasia.Similarly, the different types of tumors that may develop in the brain can produce similar MRI signals that complicate their differential diagnosis32,,,,66。To address these issues, future studies require (i) training and validation databases that balance the amount and diversity of confounding features and comorbidities (e.g., breasts with mass-like and non-mass geometries; prostates with cancer alone and combined with other prostatic pathologies; and diverse brain tumor cases), and (ii) training AI models to recognize confounding patterns (e.g., standard textural and deep learning features) to boost the performance of CADe/CADx technologies for cancer32,,,,64,,,,65,,,,66,,,,67,,,,71,,,,86,,,,87,,,,88。
Several validation, implementation, ethical, and data issues have also hindered the clinical translation of AI models for cancer detection and diagnosis.Firstly, the diagnostic performance of CADe/CADx systems still needs to be externally validated in large prospective clinical trials including diverse imaging acquisition methods, patient demographics, and reader expertise32,,,,63,,,,64,,,,65,,,,69,,,,91,,,,92。Additionally, two key obstacles to their practical clinical deployment are the lack of local data science support and the high computational cost, particularly during the training of DL models.The need for high-performance hardware and efficient algorithms demands a considerable financial investment, hindering widespread adoption in resource-constrained healthcare settings.As noted in Section 5.1, transfer learning is a promising strategy to address these computational limitations by leveraging pre-trained DL models32,,,,65,,,,68,,,,69。Another fundamental issue is the limited availability of rigorously curated data for AI model development, which may require specific labels and non-standard preprocessing that can be costly and time-consuming63。Furthermore, biases in data collection processes and unrecognized biases in clinical practices from which the training data is gathered can lead to distorted outcomes, even perpetuating societal inequalities93。Data inaccessibility due to confidentiality concerns, potential breaches compromising patient confidentiality, and strict data privacy regulations limiting collaboration also constitute important challenges63,,,,93。To address limitations in data transfer for multicenter collaborations, federated learning94is a potential strategy that relies on sharing model parameter updates rather than datasets.
AI for predicting response from MRI data
Common AI techniques for predicting response
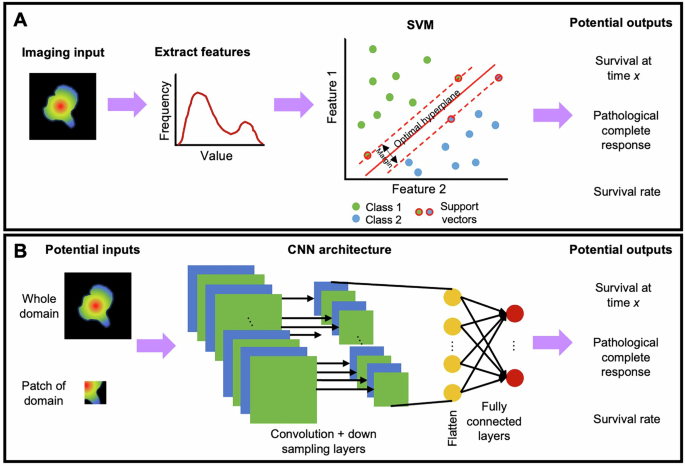
AI can be used to link imaging data to outcomes such as pathological response, time to recurrence, and overall survival.Commonly used machine learning methods for predicting response include support vector machine (SVM)95,,,,96,,,,97, regression98,,,,99, random survival forest (RSF)100, clustering101,,,,102, and CNNs99,,,,103,,,,104。Recently, more DL models105are established based on the transformer architecture or incorporating the attention mechanism106。Panels A and B of Fig.4demonstrate how SVM and CNN, respectively, can be used in prediction of response.
In Panel一个, features related to histograms of relative cerebral blood volume97or peak height of a perfusion signal107are extracted from the imaging data.Clinical information such as patient age and genetic data can also be included as features.The goal of the SVM is to takenfeatures and determine the (n-1)-dimensional hyperplanes that maximally separate (for example) patients into short, medium, or long survival, or complete response as determined by pathology.控制板bshows potential inputs to a CNN – either the whole image domain, imaging-derived features, or a patch of the domain.Extracting patches from a domain can be used to increase the amount of training data, or to reduce computational burden when working with large images.These are then input to a CNN, here represented with convolution and down sampling layers feeding into a fully connected architecture.In general, multiple sets of convolution and down-sampling layers are used.The network output accomplishes the same goal as the SVM in Panel A;namely, separating inputs into classes such as responders and non-responders, or survival at a particular time.
In brain cancer, combining clinical data with AI methods has been shown to be more effective in predicting survival outcomes than clinical methods alone107,,,,108。SVM, for instance, has been used to predict treatment outcomes for gliomas96by employing both clinical and functional features96。An SVM trained by Emblem et al.found whole tumor relative cerebral blood volume was the optimal predictor of overall survival97。CNNs have been used for the assessment and prediction of outcomes.For example, Jang et al.104distinguished between pseudo-progression and progression in patients with GBM using CNNs with long short-term memory (LSTM)109,,,,110, an ML algorithm used to train recurrent NNs.They compare two options for the CNN input data: (1) MRI (post-contrastt1-weighted images) and clinical parameters, and (2) MRI data alone.The model trained on MRI and clinical data outperformed the MRI-only model, as the AUC for predicting progression versus pseudo-progression had values of 0.83 and 0.69, respectively.This demonstrates that the combination of AI, MRI, and clinical features might help in post-treatment decision-making for patients with GBM104。
In prostate cancer, MRI-based AI has enabled the prediction of recurrence after surgery and radiotherapy111。For example, Lee et al.99developed a DL model trained on preoperative multiparametric MRI data (t2-weighted, DW-MRI, and DCE-MRI) to predict long-term post-surgery recurrence-free survival.Using Cox models and Kaplan-Meier survival analysis, the features obtained from multiparametric MRI data via their DL model outperformed clinical and radiomics features, while the combination of clinical and DL features yielded the best predictive performance.Additionally, in breast cancer, prognostic CNN models trained on MRI data have also enabled the prediction of pathological complete response for neoadjuvant chemotherapy112。
Current clinical needs
Common clinical challenges in oncology include the stratification of patients by treatment response, risk of relapse, and overall survival113,,,,114。Identification of patient outcomes before treatment begins (or early during treatment) could help select, escalate, or de-escalate prescribed treatments, as well as tailor personalized follow-up schedules.It is important to note that analyses (AI-based and otherwise) leveraging multi-parametric MRI as compared to biopsy-based stratification also enable longitudinal whole-tumor coverage addressing the sampling bias limitations of biopsy113。Furthermore, many of these AI-based approaches leveraging imaging data achieve greater performance over standard clinical information (e.g., age, sex, extent of surgery) alone113,,,,114。
Barriers to practical deployment in the clinical setting
Although AI-based approaches have demonstrated greater performance over standard clinical information in some settings, these studies are typically done in specific cohorts with limited external validation and therefore have limited generalizability115。The variability in MR acquisition techniques between manufacturers, scanner types, protocols, and institutions can lead to substantial bias in training image-guided AIs without proper data harmonization116, such as that completed by Marzi et al.117, who were able to reduce site effects after data harmonization in a study of T1-weighted MRI data from 1740 healthy subjects at 36 sites using a harmonizer transformer as part of the preprocessing steps for a machine learning pipeline.Moreover, restrictions on inter-institution sharing of patient data118(due to concerns such as privacy) may restrict the verification of model generalizability.These limitations, alongside uncertainty in the AI models themselves, create substantial barriers to the widespread clinical adoption of AI models for response prediction.Federated learning could be a realistic strategy to bypass the complexity of inter-institutional data sharing.Instead of requiring assembling data from different institutions into a centralized large-scale dataset, federated learning enables training of the AI model on decentralized, private datasets in multiple sites independently, then only the trained parameters (instead of original data) will be shared between training sites to generate the globally tuned model119,,,,120
Efforts to construct large, publicly available MRI datasets for brain tumors are ongoing.However, careful considerations need to be made while using them in training, testing or validating AI-based model121。One significant issue is the potential for overlaps between different datasets (i.e., multiple datasets containing the same patient), which can reduce the number of uniquely available data.More specifically, the IvyGAP Radiomics dataset contains the pre-operative MRIs of the IvyGap dataset with additional segmentations and derived radionics parameters.Moreover, the BraTS 2021 dataset contains data that was available in the previous BraTS challenges and other public datasets such as the TCGA-LGG (65 patients), TCGA GBM (102 patients), and Ivy Gap (30 patients).This may lead to redundancy and diminish the overall diversity of the training data121。Furthermore, these datasets have been published for more than a decade and some of them have undergone updates, adding a higher variability in protocols, scans quality, and evolving WHO classification.We identify similar issues in prostate cancer122, particularly when it comes to the age of the publicly available datasets (many being 10+ years old) and overlap between datasets (which may or may not be known between sets).Dataset sizes are also often small, especially once missing or low-quality data is removed.
Efforts for breast cancer data standardization are also in progress.For instance, Kilintzis et al.123attempted to produce a harmonized dataset from 5 publicly available datasets from the TCIA platform, including 2035 patients.More generally, Kosvyra et al.124propose a new methodology for assessing the data quality of cancer imaging repositories by incorporating a three-step procedure that includes a data integration quality check tool to ensure compliance with quality requirements.
Federated learning could be a practical strategy to overcome the complexity of inter-institutional data sharing.Instead of assembling data from different institutions into a centralized large-scale dataset, federated learning enables the training of the AI model on decentralized, private datasets in multiple sites independently, then only the trained parameters (instead of original data) are shared between training sites to generate the globally tuned model.We discuss these points via the results presented in Rauniyar et al.119and Guan et al.120。
Beyond data variation, the cancer population itself is heterogeneous (as categorized by the cancer subtypes, staging, patient demographics, etc.), which leads to differences in therapy response between patients125。Moreover, novel therapies can be introduced into clinical care, the therapeutic regimens vary between institutions, and treatments can be refined, leading to significant differences in datasets collected at different times126。Overall, the trade-off between the sample size and homogeneity of accessible datasets to train AI models is a major barrier to the robust application of AI for response prediction.One method that could help combat generalizability issues with small datasets is pretraining.For example, Yuan et al.127and Han et al.128pretrain convolutional neural networks using the ImageNet129dataset to successfully classify risk in prostate tumors and differentiate between long- and short-term glioma survivors, respectively.As an alternative to pretraining on ImageNet, Wen et al.130explore the possible benefits of pretraining using medical images.While their study ultimately found that ImageNet pretraining provided more accurate results, medical pretraining showed potential.As such, it could be useful to construct a large medical image database like ImageNet to further explore pretraining for medical problems using medical images.张等。131explore another possible workflow for fine-tuning a network designed to classify breast cancer molecular subtypes from DCE-MRI data.Rather than relying on a separate, large dataset for pre-training, they separate their data into a training set and two testing sets – A and B. They then compare (1) testing the network with both A and B, (2) fine-tuning with B and testing with A, and (3) fine-tuning with A and testing with B. They conclude that finetuning with this method increases accuracy.Thus, in a situation where the dataset of interest is very small, initial network training could be completed on large, publicly available datasets, then fine-tuning and testing could be completed using the dataset of interest.
Many AI-based algorithms lack interpretability, thereby also limiting their clinical adoption.Furthermore, most AI-based approaches for outcome prediction are deployed with set parameters that govern their sensitivity and specificity.The patient-specific use of these models may require their optimization to fit the specific wishes of the patient, their family, and their clinical team.This patient-specific optimization creates an additional barrier to clinical deployment.The integration of AI-based prediction algorithms into the clinical workflow faces additional challenges as it has not been well established and currently generates additional work for the clinical team.This limits their adoption to clinics that have a robust computational infrastructure, clinicians who are willing to spend extra time generating the data, and ready access to the necessary data for the algorithm.
讨论
AI-based methods have established some measure of success in several areas of MRI including image acquisition, reconstruction, registration, and segmentation, as well as assisting in diagnosis and prognosis.Some registration and segmentation techniques have even been approved by the FDA for clinical application69,,,,91,,,,92,,,,132。However, while the application of AI techniques for MRI in cancer clearly has tremendous potential, three major challenges persist: (1) model generalizability, (2) model interpretability, and (3) establishing confidence in the output of an AI model.Indeed, these problems are common to many applications of AI in medicine.
The limited progress in being able to transport an AI method from one institute to another, and from one disease setting to another (i.e., generalizability), is heavily influenced by both the data and the devices employed to capture the data133。The wide variety of the available scanners (both from different manufacturers and then different models with a manufacturer), as well as the protocols they run, can further hamper generalizability as the data employed for training may not adequately sample all the acquisition scenarios encountered in the testing data set133,,,,134。The lack of standardization in quality assurance and control (QA/QC) adds additional complexities135。Even when an AI method has had success in one clinical setting, it does not guarantee it will be successful when applied in another disease setting due to variations in patient characteristics and previously received treatments136。Thus, retraining the AI model is required and this can introduce nontrivial changes to the AI architecture or method of implementation.This is especially true in cancer, where the differences between diseases and the site of origin can introduce tremendous anatomical and physiological heterogeneities that may confound a previously trained AI model.It is increasingly recognized that each model should be evaluated with local data in the disease setting for which it is intended to be used137。
A fundamental issue with the majority of AI-based analysis is their limited “interpretability†in the sense of only providing a limited understanding of the relationships between model input and model output;that is, there is limited insight into how the AI model gets from cause to effect.This is an active area of investigation72,,,,138,,,,139and is now well-recognized for being extremely important in domains that have high-consequence decisions like oncology140,,,,141。For example, lack of interpretability generates problems when trying to identify optimal interventions for a particular patient where it is critical to understand why an AI model selects a particular therapeutic regimen over another.In particular, for an AI model built on population-based MRI data, does the training data set adequately capture the unique characteristics of the features in the individual’s MRI data?One attractive way forward is to link mechanism-based modeling with AI methods and scientific machine learning in which known/established biological and physical laws can be explicitly incorporated in the AI algorithm142,,,,143,,,,144。This can have the additional benefit of increasing confidence in the method.
The challenge in establishing confidence is not only related to the lack of model generalizability and interpretability, but also concern over the ethical issues associated with applying AI techniques to healthcare data as well as maintaining patient privacy and data security.This is particularly true with medical imaging data in which detailed anatomical features can be rendered in 3D.Importantly, once introduced in the clinical workflow, the performance of the AI tool must be continuously monitored as the data inputs are adjusted with new imaging hardware, software, and methods of image acquisition and subsequent analysis.More generally, the Office of Science and Technology Policy has published a white paper entitled, “A Blueprint for an AI Bill of Rightsâ€145designed to guide the safe, effective, and unbiased development and application of AI.
Beyond the issues related to generalizability, interpretability, and confidence enumerated above, there may be fundamental limitations to what AI can contribute to cancer imaging.For example, it is important to note that every AI-based method requires a training set and since cancer is notoriously heterogeneous across both space and time, there are fundamental limits to what a population-based method can achieve.Indeed, this problem manifests itself in everything from image reconstruction to predicting response if the specific details of the pathology under investigation are not well-characterized in the training set.For a disease as heterogeneous as cancer, and an imaging modality as flexible as MRI, this seems to be a difficult problem to overcome with an AI-only approach.
结论
AI has shown promise in accelerating image acquisition and reconstruction methods to maintain high spatial resolution in a fraction of the time typically required to obtain such an image.It has also been a useful tool for improving image segmentation and registration, as well as offering valuable results in both diagnostic and prognostic settings.However, questions remain about the interpretability and generalizability of these techniques, especially when a method is ported from one disease setting to another or, even, from one institution to another institution.The dearth of robust QA/QC and ethics-ensuring methods means that much work is left to be done to maintain patient safety given the current status of AI techniques in cancer imaging and healthcare.Finally, questions concerning fundamental limitations for any method that requires a large training dataset remain.
数据可用性
No datasets were generated or analyzed during the current study.
参考
Chen, Y. et al.AI-based reconstruction for Fast MRI—A systematic review and meta-analysis.Proc。IEEE 110, 224–245 (2022).
文章一个 CAS一个 Google Scholar一个
Shimron, E. & Perlman, O. AI in MRI: Computational frameworks for a faster, optimized, and automated imaging workflow.生物工程 10, 492 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Chen,W。等。Artificial intelligence powered advancements in upper extremity joint MRI: A review.Heliyon 10, e28731 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Li,C。等。Artificial intelligence in multiparametric magnetic resonance imaging: A review.医学物理。 49,,,,https://doi.org/10.1002/mp.15936(2022).
McCarthy, J., Minsky, M. L., Rochester, N. & Shannon, C. E. A proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955.AI Mag. 27, 12 (2006).
Ongie, G. et al.Deep learning techniques for inverse problems in imaging.IEEE J. SEL。Areas Inf.理论 1, 39–56 (2020).
文章一个 Google Scholar一个
Shen, D., Wu, G. & Suk, H. I. Deep learning in medical image analysis.安努。Rev. Biomed.工程。 19, 221–248 (2017).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lundervold, A. S. & Lundervold, A. An overview of deep learning in medical imaging focusing on MRI.Z. F. ür.Med Phys。 29, 102–127 (2019).
文章一个 Google Scholar一个
Karimi, D., Dou, H., Warfield, S. K. & Gholipour, A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis.医学图像肛门。 65, 101759 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Iglesias, J. E. et al.SynthSR: A public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry.科学。ADV。 9, eadd3607 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
De Leeuw Den Bouter, M. L. et al.Deep learning-based single image super-resolution for low-field MR brain images.科学。代表。 12, 6362 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Guan, H. & Liu, M. Domain adaptation for medical image analysis: a survey.IEEE Trans。生物。工程。 69, 1173–1185 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Tamir, J. I. et al.Computational MRI with physics-based constraints: application to multicontrast and quantitative imaging.IEEE Signal Process Mag. 37, 94–104 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Dang, H. N. et al.MRâ€zero meets RARE MRI: Joint optimization of refocusing flip angles and neural networks to minimizet 2â€induced blurring in spin echo sequences.Magn.Reson Med 90, 1345–1362 (2023).
文章一个 PubMed一个 Google Scholar一个
Loktyushin, A. et al.MRzero - Automated discovery of MRI sequences using supervised learning.Magn.Reson Med 86, 709–724 (2021).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Aggarwal, H. K. & Jacob, M. J-MoDL: Joint model-based deep learning for optimized sampling and reconstruction.IEEE J. SEL。顶部。Signal Process 14, 1151–1162 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Bahadir C. D., Dalca A. V. & Sabuncu M. R. Learning-Based Optimization of the Under-Sampling Pattern in MRI.In: Chung A. C. S., Gee J. C., Yushkevich P. A., Bao S., eds.Information Processing in Medical Imaging。Vol 11492. Lecture Notes in Computer Science.Springer International Publishing;780–792.(2019)。
Ravula S., Levac B., Jalal A., Tamir J. I. & Dimakis A. G. Optimizing Sampling Patterns for Compressed Sensing MRI with Diffusion Generative Models.在线发布。https://doi.org/10.48550/ARXIV.2306.03284(2023)。
Wang, G., Luo, T., Nielsen, J. F., Noll, D. C. & Fessler, J. A. B-spline parameterized joint optimization of reconstruction and K-Space Trajectories (BJORK) for accelerated 2D MRI.IEEE Trans。医学成像 41, 2318–2330 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lustig, M., Donoho, D. & Pauly, J. M. Sparse MRI: The application of compressed sensing for rapid MR imaging.Magn.reson。医学 58, 1182–1195 (2007).
文章一个 PubMed一个 Google Scholar一个
Aggarwal, H. K., Mani, M. P. & Jacob, M. MoDL: Model-based deep learning architecture for inverse problems.IEEE Trans。医学成像 38, 394–405 (2019).
文章一个 PubMed一个 Google Scholar一个
Hammernik, K. et al.Learning a variational network for reconstruction of accelerated MRI data.Magn.reson。医学 79, 3055–3071 (2018).
文章一个 PubMed一个 Google Scholar一个
Jalal A. et al.Robust Compressed Sensing MRI with Deep Generative Priors.In: Ranzato M., Beygelzimer A., Dauphin Y., Liang P. S., Vaughan J. W., eds.神经信息处理系统的进步。Vol 34. Curran Associates, Inc.;14938–14954.https://proceedings.neurips.cc/paper_files/paper/2021/file/7d6044e95a16761171b130dcb476a43e-Paper.pdf。(2021)。
Luo, G., Blumenthal, M., Heide, M. & Uecker, M. Bayesian MRI reconstruction with joint uncertainty estimation using diffusion models.Magn.reson。医学 90, 295–311 (2023).
文章一个 PubMed一个 Google Scholar一个
Johnson, P. M. et al.Deep learning reconstruction enables prospectively accelerated clinical knee MRI.放射学 307, e220425 (2023).
文章一个 PubMed一个 Google Scholar一个
Chen,F。等。Variable-density single-shot fast spin-echo MRI with deep learning reconstruction by using variational networks.放射学 289, 366–373 (2018).
文章一个 PubMed一个 Google Scholar一个
Yankeelov, T. & Gore, J. Dynamic contrast-enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples.Curr。医学Imaging Rev. 3, 91–107 (2007).
文章一个 CAS一个 Google Scholar一个
Gong, E., Pauly, J. M., Wintermark, M. & Zaharchuk, G. Deep learning enables reduced gadolinium dose for contrastâ€enhanced brain MRI.J. Magn。reson。成像 48, 330–340 (2018).
文章一个 PubMed一个 Google Scholar一个
Muckley, M. J. et al.Results of the 2020 fastMRI challenge for machine learning MR image reconstruction.IEEE Trans。医学成像 40, 2306–2317 (2021).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Daye, D. et al.Implementation of clinical artificial intelligence in radiology: who decides and how?放射学 305, 555–563 (2022).
文章一个 PubMed一个 Google Scholar一个
Hoebel, K. V. et al.Expert-centered evaluation of deep learning algorithms for brain tumor segmentation.收音机。Artif.Intell。 6, e220231 (2024).
文章一个 Google Scholar一个
Cè, M. et al.Artificial Intelligence in brain tumor imaging: a step toward personalized medicine.Curr。Oncol。 30, 2673–2701 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Egger, J. et al.GBM Volumetry using the 3D Slicer Medical Image Computing Platform.科学。代表。 3, 1364 (2013).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lambert, S. I. et al.关于医疗保健专业人员在医院中接受人工智能的综合综述。Npj Digit.医学 6, 111 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Baroudi, H. et al.Automated contouring and planning in radiation therapy: what is ‘Clinically Acceptable’?诊断 13, 667 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Pal, R. et al.Lumbar Spine Tumor Segmentation and Localization in T2 MRI Images Using AI.在线发布。https://doi.org/10.48550/ARXIV.2405.04023(2024)。
Kundal, K., Rao, K. V., Majumdar, A., Kumar, N. & Kumar, R. Comprehensive benchmarking of CNN-based tumor segmentation methods using multimodal MRI data.计算。生物。医学 178, 108799 (2024).
文章一个 PubMed一个 Google Scholar一个
Eppenhof, K. A. J. & Pluim, J. P. W. Pulmonary CT registration through supervised learning with convolutional neural networks.IEEE Trans。医学成像 38, 1097–1105 (2019).
文章一个 PubMed一个 Google Scholar一个
Osman, A. F. I., Al-Mugren, K. S., Tamam, N. M. & Shahine, B. Deformable registration of magnetic resonance images using unsupervised deep learning in neuro-/radiation oncology.辐射。Oncol。 19, 61 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Rudie, J. D. et al.Longitudinal assessment of posttreatment diffuse glioma tissue volumes with three-dimensional convolutional neural networks.收音机。Artif.Intell。 4, e210243 (2022).
文章一个 Google Scholar一个
Teuwen, J., Gouw, Z. A. R. & Sonke, J. J. Artificial intelligence for image registration in radiation oncology.Semin Radiat.Oncol。 32, 330–342 (2022).
文章一个 PubMed一个 Google Scholar一个
Simonovsky M., Gutiérrez-Becker B., Mateus D., Navab N. & Komodakis N. A Deep Metric for Multimodal Registration.In: Ourselin S., Joskowicz L., Sabuncu M. R., Unal G., Wells W., eds.Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016。Vol 9902. Lecture Notes in Computer Science.Springer International Publishing 10–18 (2016).
Haskins, G. et al.Learning deep similarity metric for 3D MR–TRUS image registration.int。J. Comput。协助。收音机。外科。 14, 417–425 (2019).
文章一个 Google Scholar一个
Mohseni Salehi, S. S., Khan, S., Erdogmus, D. & Gholipour, A. Real-time deep pose estimation with geodesic loss for image-to-template rigid registration.IEEE Trans。Med Imaging 38, 470–481 (2019).
文章一个 PubMed一个 Google Scholar一个
Estienne, T. et al.Deep learning-based concurrent brain registration and tumor segmentation.正面。计算。Neurosci。 14, 17 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Bakas S. et al.Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge.Published online November 5.https://doi.org/10.17863/CAM.38755(2018)。
Balakrishnan, G., Zhao, A., Sabuncu, M. R., Guttag, J. & Dalca, A. V. VoxelMorph: A Learning Framework for Deformable Medical Image Registration.IEEE Trans。医学成像 38, 1788–1800 (2019).
文章一个 Google Scholar一个
Dalca, A. V., Balakrishnan, G., Guttag, J. & Sabuncu, M. R. Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces.医学图像肛门。 57, 226–236 (2019).
文章一个 PubMed一个 Google Scholar一个
Pellicer-Valero, O. J. et al.Deep learning for fully automatic detection, segmentation, and Gleason grade estimation of prostate cancer in multiparametric magnetic resonance images.科学。代表。 12, 2975 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Cardenas, C. E., Yang, J., Anderson, B. M., Court, L. E. & Brock, K. B. Advances in auto-segmentation.Semin Radiat.Oncol。 29, 185–197 (2019).
文章一个 PubMed一个 Google Scholar一个
Warfield, S. K., Zou, K. H. & Wells, W. M. Validation of image segmentation by estimating rater bias and variance.哲学反式。R. Soc。数学。物理。工程。科学。 366, 2361–2375 (2008).
Ramesh, K. K. D., Kumar, G. K., Swapna, K., Datta, D. & Rajest, S. S. A Review of Medical Image Segmentation Algorithms.EAI Endorsed Trans.Pervasive Health Technol. 7, e6 (2021).
文章一个 Google Scholar一个
Jungo A. et al.On the Effect of Inter-observer Variability for a Reliable Estimation of Uncertainty of Medical Image Segmentation.In: Frangi A. F., Schnabel J. A., Davatzikos C., Alberola-López C., Fichtinger G., eds.Medical Image Computing and Computer Assisted Intervention – MICCAI 2018。Vol 11070. Lecture Notes in Computer Science.Springer International Publishing;682–690.(2018)。
Renard, F., Guedria, S., Palma, N. D. & Vuillerme, N. Variability and reproducibility in deep learning for medical image segmentation.科学。代表。 10, 13724 (2020).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Tixier, F., Um, H., Young, R. J. & Veeraraghavan, H. Reliability of tumor segmentation in glioblastoma: Impact on the robustness of MRIâ€radiomic features.医学物理。 46, 3582–3591 (2019).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Poirot, M. G. et al.Robustness of radiomics to variations in segmentation methods in multimodal brain MRI.科学。代表。 12, 16712 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Koçak, B. Key concepts, common pitfalls, and best practices in artificial intelligence and machine learning: focus on radiomics.诊断。间。收音机。 28, 450–462 (2022).
文章一个 Google Scholar一个
Kocak, B., Durmaz, E. S., Ates, E. & Kilickesmez, O. Radiomics with artificial intelligence: a practical guide for beginners.诊断。间。收音机。 25, 485–495 (2019).
文章一个 Google Scholar一个
Chen,Z。等。What matters in radiological image segmentation?Effect of segmentation errors on the diagnostic related features.J. Digit Imaging 36, 2088–2099 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Horvat, N., Papanikolaou, N. & Koh, D. M. Radiomics beyond the hype: a critical evaluation toward oncologic clinical use.收音机。Artif.Intell。 6, e230437 (2024).
文章一个 Google Scholar一个
Fu, Y. et al.Deep learning in medical image registration: a review.物理。医学生物。 65, 20TR01 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
MÃ¥rtensson, G. et al.The reliability of a deep learning model in clinical out-of-distribution MRI data: A multicohort study.医学图像肛门。 66, 101714 (2020).
文章一个 PubMed一个 Google Scholar一个
Bi, W. L. et al.Artificial intelligence in cancer imaging: Clinical challenges and applications.CA Cancer J. Clin。 69, 127–157 (2019).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wildeboer, R. R., Van Sloun, R. J. G., Wijkstra, H. & Mischi, M. Artificial intelligence in multiparametric prostate cancer imaging with focus on deep-learning methods.计算。Methods Prog.生物。 189, 105316 (2020).
文章一个 Google Scholar一个
Jones, M. A., Islam, W., Faiz, R., Chen, X. & Zheng, B. Applying artificial intelligence technology to assist with breast cancer diagnosis and prognosis prediction.正面。Oncol。 12, 980793 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Chakrabarty, S. et al.MRI-based identification and classification of major intracranial tumor types by using a 3D Convolutional Neural Network: A retrospective multi-institutional analysis.收音机。Artif.Intell。 3, e200301 (2021).
文章一个 Google Scholar一个
Saha, A., Hosseinzadeh, M. & Huisman, H. End-to-end prostate cancer detection in bpMRI via 3D CNNs: Effects of attention mechanisms, clinical priori and decoupled false positive reduction.医学图像肛门。 73, 102155 (2021).
文章一个 PubMed一个 Google Scholar一个
Giger, M. L. Machine learning in medical imaging.J. Am。科尔。收音机。 15, 512–520 (2018).
文章一个 Google Scholar一个
Twilt, J. J., Van Leeuwen, K. G., Huisman, H. J., Fütterer, J. J. & De Rooij, M. Artificial intelligence based algorithms for prostate cancer classification and detection on magnetic resonance imaging: a narrative review.诊断 11, 959 (2021).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Antropova, N., Huynh, B. Q. & Giger, M. L. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets.医学物理。 44, 5162–5171 (2017).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sardanelli, F. et al.The paradox of MRI for breast cancer screening: high-risk and dense breasts—available evidence and current practice.Insights Imaging 15, 96 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead.纳特。马赫。Intell。 1, 206–215 (2019).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Hatherley, J., Sparrow, R. & Howard, M. The virtues of interpretable medical AI.Camb.Q Health Ethics 33, 323–332 (2024).
文章一个 Google Scholar一个
Salahuddin, Z., Woodruff, H. C., Chatterjee, A. & Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods.计算。生物。医学 140, 105111 (2022).
文章一个 PubMed一个 Google Scholar一个
Band, S. et al.Application of explainable artificial intelligence in medical health: A systematic review of interpretability methods.inf。医学解锁 40, 101286 (2023).
文章一个 Google Scholar一个
Champendal, M., Müller, H., Prior, J. O. & Dos Reis, C. S. A scoping review of interpretability and explainability concerning artificial intelligence methods in medical imaging.欧元。J. Radio. 169, 111159 (2023).
文章一个 Google Scholar一个
Lipkova, J. et al.Artificial intelligence for multimodal data integration in oncology.癌细胞 40, 1095–1110 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Paverd, H., Zormpas-Petridis, K., Clayton, H., Burge, S. & Crispin-Ortuzar, M. Radiology and multi-scale data integration for precision oncology.Npj Precis.Oncol。 8, 158 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Metzcar, J., Jutzeler, C. R., Macklin, P., Köhn-Luque, A. & Brüningk, S. C. A review of mechanistic learning in mathematical oncology.正面。免疫。 15, 1363144 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lorenzo, G. et al.Patient-specific, mechanistic models of tumor growth incorporating artificial intelligence and big data.安努。Rev. Biomed.工程。 26, 529–560 (2024).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Bosque, J. J. et al.Metabolic activity grows in human cancers pushed by phenotypic variability.Iscience 26, 106118 (2023).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Raissi, M., Perdikaris, P. & Karniadakis, G. E. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations.J. Comput。物理。 378, 686–707 (2019).
文章一个 Google Scholar一个
Zhang, R. Z. et al.Personalized predictions of Glioblastoma infiltration: Mathematical models, Physics-Informed Neural Networks and multimodal scans.医学图像肛门。 101, 103423 (2025).
文章一个 PubMed一个 Google Scholar一个
Elmarakeby, H. A. et al.Biologically informed deep neural network for prostate cancer discovery.自然 598, 348–352 (2021).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lagergren, J. H., Nardini, J. T., Baker, R. E., Simpson, M. J. & Flores, K. B. Biologically-informed neural networks guide mechanistic modeling from sparse experimental data.PLOS计算。生物。 16, e1008462 (2020).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Witowski, J. et al.Improving breast cancer diagnostics with deep learning for MRI.科学。翻译。医学 14, eabo4802 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Meyerâ€Base, A. et al.人工智能-enhanced diagnosis of challenging lesions in breast MRI: a methodology and application primer.J. Magn。reson。成像 54, 686–702 (2021).
文章一个 PubMed一个 Google Scholar一个
Rosenkrantz, A. B. & Taneja, S. S. Radiologist, be aware: ten pitfalls that confound the interpretation of multiparametric prostate MRI.是。J. Roentgenol. 202, 109–120 (2014).
文章一个 Google Scholar一个
Cao, R. et al.Joint prostate cancer detection and Gleason score prediction in mp-MRI via FocalNet.IEEE Trans。医学成像 38, 2496–2506 (2019).
文章一个 PubMed一个 Google Scholar一个
Bashkanov, O. et al.Automatic detection of prostate cancer grades and chronic prostatitis in biparametric MRI.计算。Methods Prog.生物。 239, 107624 (2023).
文章一个 Google Scholar一个
Wang, J. Y. et al.Stratified assessment of an FDA-cleared deep learning algorithm for automated detection and contouring of metastatic brain tumors in stereotactic radiosurgery.辐射。Oncol。 18, 61 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Jiang, Y., Edwards, A. V. & Newstead, G. M. Artificial Intelligence applied to breast MRI for improved diagnosis.放射学 298, 38–46 (2021).
文章一个 PubMed一个 Google Scholar一个
Khan, B. et al.Drawbacks of artificial intelligence and their potential solutions in the healthcare sector.生物。母校。设备 1, 731–738 (2023).
文章一个 Google Scholar一个
Pati, S. et al.Federated learning enables big data for rare cancer boundary detection.纳特。社区。 13, 7346 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Cortes, C. & Vapnik, V. Support-vector networks.马赫。学习。 20, 273–297 (1995).
文章一个 Google Scholar一个
Gutman, D. A. et al.MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA Glioblastoma data set.放射学 267, 560–569 (2013).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Emblem, K. E. et al.A generic support vector machine model for Preoperative Glioma Survival Associations.放射学 275, 228–234 (2015).
文章一个 PubMed一个 Google Scholar一个
Cox, D. R. Regression models and life-tables.J. R. Stat。Soc。ser。B Stat.methodol。 34, 187–202 (1972).
文章一个 Google Scholar一个
Lee, H. W. et al.Novel multiparametric magnetic resonance imaging-based deep learning and clinical parameter integration for the prediction of long-term biochemical recurrence-free survival in prostate cancer after Radical Prostatectomy.Cancers 15, 3416 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ishwaran H., Kogalur U. B., Blackstone E. H. & Lauer M. S. Random survival forests.安。应用。统计。2,,,,https://doi.org/10.1214/08-AOAS169(2008)。Xu, R. & Wunsch, D. C. Clustering algorithms in biomedical research: a review.
IEEE Rev. Biomed.工程。 3, 120–154 (2010).
文章一个 PubMed一个 Google Scholar一个
Andersen, E. K. F., Kristensen, G. B., Lyng, H. & Malinen, E. Pharmacokinetic analysis and k-means clustering of DCEMR images for radiotherapy outcome prediction of advanced cervical cancers.Acta Oncol。 50, 859–865 (2011).
文章一个 PubMed一个 Google Scholar一个
Anwar, S. M. et al.Medical image analysis using convolutional neural networks: a review.J. Med。系统。 42, 226 (2018).
文章一个 PubMed一个 Google Scholar一个
Jang, B. S., Jeon, S. H., Kim, I. H. & Kim, I. A. Prediction of Pseudoprogression versus progression using machine learning algorithm in Glioblastoma.科学。代表。 8, 12516 (2018).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zhou, X. et al.Attention mechanism based multi-sequence MRI fusion improves prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer.辐射。Oncol。 18, 175 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Vaswani A., et al.Attention is All you Need.In: Guyon I., Luxburg U. V., Bengio S., et al., eds.神经信息处理系统的进步。Vol 30. Curran Associates, Inc.;https://proceedings.neurips.cc/paper_files/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf。(2017)。
Macyszyn, L. et al.Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques.Neuro-Oncol. 18, 417–425 (2016).
文章一个 PubMed一个 Google Scholar一个
Kickingereder, P. et al.Radiomic profiling of Glioblastoma: Identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models.放射学 280, 880–889 (2016).
文章一个 PubMed一个 Google Scholar一个
Hochreiter,S。&Schmidhuber,J。长期记忆。神经计算。 9, 1735–1780 (1997).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Van Houdt, G., Mosquera, C. & Nápoles, G. A review on the long short-term memory model.Artif.Intell。修订版 53, 5929–5955 (2020).
文章一个 Google Scholar一个
Li, H. et al.Machine learning in prostate MRI for prostate cancer: current status and future opportunities.诊断 12, 289 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Khan, N., Adam, R., Huang, P., Maldjian, T. & Duong, T. Q. Deep learning prediction of pathologic complete response in breast cancer using MRI and other clinical data: a systematic review.断层扫描 8, 2784–2795 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Cè M., et al.Artificial intelligence in breast cancer imaging: risk stratification, lesion detection and classification, treatment planning and prognosis—a narrative review.Explor Target Anti-Tumor Ther。Published online December27, 795–816.(2022)
朱,M。等。Artificial intelligence in the radiomic analysis of glioblastomas: A review, taxonomy, and perspective.前Oncol。 12, 924245 (2022).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zhang, B., Shi, H. & Wang, H. Machine learning and AI in cancer prognosis, prediction, and treatment selection: a critical approach.J. Multidiscip.健康 16, 1779–1791 (2023).
文章一个 Google Scholar一个
Papadimitroulas, P. et al.Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization.物理。医学 83, 108–121 (2021).
文章一个 PubMed一个 Google Scholar一个
Marzi, C. et al.Efficacy of MRI data harmonization in the age of machine learning: a multicenter study across 36 datasets.科学。数据 11, 115 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Basu, K., Sinha, R., Ong, A. & Basu, T. Artificial intelligence: How is it changing medical sciences and its future?Indian J. Dermatol 65, 365 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Rauniyar, A. et al.Federated learning for medical applications: a taxonomy, current trends, challenges, and future research directions.IEEE互联网事物。 11, 7374–7398 (2024).
文章一个 Google Scholar一个
Guan, H., Yap, P. T., Bozoki, A. & Liu, M. Federated learning for medical image analysis: A survey.Pattern Recognit. 151, 110424 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Andaloussi M. A., Maser R., Hertel F., Lamoline F. & Husch A. D. Exploring Adult Glioma through MRI: A Review of Publicly Available Datasets to Guide Efficient Image Analysis.在线发布。https://doi.org/10.48550/ARXIV.2409.00109(2024)。
Sunoqrot, M. R. S., Saha, A., Hosseinzadeh, M., Elschot, M. & Huisman, H. Artificial intelligence for prostate MRI: open datasets, available applications, and grand challenges.欧元。收音机。经验。 6, 35 (2022).
文章一个 Google Scholar一个
Kilintzis, V. et al.Public data homogenization for AI model development in breast cancer.欧元。收音机。经验。 8, 42 (2024).
文章一个 Google Scholar一个
Kosvyra, A., Filos, D. T., Fotopoulos, D. T. H., Tsave, O. & Chouvarda, I. Toward ensuring data quality in multi-site cancer imaging repositories.信息 15, 533 (2024).
文章一个 Google Scholar一个
Bedard, P. L., Hansen, A. R., Ratain, M. J. & Siu, L. L. Tumour heterogeneity in the clinic.自然 501, 355–364 (2013).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Booth, C. M., Karim, S. & Mackillop, W. J. Real-world data: towards achieving the achievable in cancer care.纳特。Rev. Clin.Oncol。 16, 312–325 (2019).
文章一个 PubMed一个 Google Scholar一个
Yuan, Y. et al.Prostate cancer classification with multiparametric MRI transfer learning model.医学物理。 46, 756–765 (2019).
文章一个 PubMed一个 Google Scholar一个
Han, W. et al.Deep transfer learning and radiomics feature prediction of survival of patients with high-grade Gliomas.是。J. Neuroradiol。 41, 40–48 (2020).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Deng J., et al.ImageNet: A large-scale hierarchical image database.在:2009 IEEE Conference on Computer Vision and Pattern Recognition。IEEE;248–255 (2009).
Wen, Y., Chen, L., Deng, Y. & Zhou, C. Rethinking pre-training on medical imaging.J. Vis。社区。图像代表。 78, 103145 (2021).
文章一个 Google Scholar一个
Zhang,Y。等。Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers.欧元。收音机。 31, 2559–2567 (2021).
文章一个 Google Scholar一个
美国食品和药物管理局。Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices。https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices。(2024)。
Eche, T., Schwartz, L. H., Mokrane, F. Z. & Dercle, L. Toward generalizability in the deployment of artificial intelligence in radiology: role of computation stress testing to overcome underspecification.收音机。Artif.Intell。 3, e210097 (2021).
文章一个 Google Scholar一个
Futoma, J., Simons, M., Panch, T., Doshi-Velez, F. & Celi, L. A. The myth of generalisability in clinical research and machine learning in health care.Lancet Digit Health 2, e489–e492 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Adjeiwaah, M., Garpebring, A. & Nyholm, T. Sensitivity analysis of different quality assurance methods for magnetic resonance imaging in radiotherapy.物理。Imaging Radiat.Oncol。 13, 21–27 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Yang Y., Zhang H., Gichoya J. W., Katabi D. & Ghassemi, M. The limits of fair medical imaging AI in real-world generalization.纳特。医学。Published online June 28,https://doi.org/10.1038/s41591-024-03113-4(2024)。
Albahri, A. S. et al.A systematic review of trustworthy and explainable artificial intelligence in healthcare: Assessment of quality, bias risk, and data fusion.inf。融合 96, 156–191 (2023).
文章一个 Google Scholar一个
Linardatos, P., Papastefanopoulos, V. & Kotsiantis, S. Explainable AI: A review of machine learning interpretability methods.熵 23, 18 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Murdoch, W. J., Singh, C., Kumbier, K., Abbasi-Asl, R. & Yu, B. Definitions, methods, and applications in interpretable machine learning.Proc。纳特。学院。科学。 116, 22071–22080 (2019).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Banegas-Luna, A. J. et al.Towards the interpretability of machine learning predictions for medical applications targeting personalised therapies: a cancer case survey.int。J. Mol。科学。 22, 4394 (2021).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Kuenzi, B. M. et al.Predicting drug response and synergy using a deep learning model of human cancer cells.癌细胞 38, 672–684.e6 (2020).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wysocka, M., Wysocki, O., Zufferey, M., Landers, D. & Freitas, A. A systematic review of biologically-informed deep learning models for cancer: fundamental trends for encoding and interpreting oncology data.BMC Bioinforma. 24, 198 (2023).
文章一个 Google Scholar一个
Cuomo, S. et al.Scientific machine learning through physics–informed neural networks: where we are and what’s next.J. Sci。Comput 92, 88 (2022).
文章一个 Google Scholar一个
Jiang,Y。等。Biology-guided deep learning predicts prognosis and cancer immunotherapy response.纳特。社区。 14, 5135 (2023).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
White House Office of Science and Technology Policy.Blueprint for an AI Bill of Rights。https://www.whitehouse.gov/ostp/ai-bill-of-rights/(2022).
致谢
We thank the National Cancer Institute for funding through 1R01CA260003, U24CA226110, U01CA253540, and P30CA016672.We thank the Cancer Prevention and Research Institute of Texas for funding through RP220225.We thank the National Science Foundation for funding through CCF-2239687 (CAREER), IFML 2019844, AF 1901292. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DE2137420.This project was supported by the Resources of the Image Guided Cancer Therapy Research Program at The University of Texas MD Anderson Cancer Center as well as the Joint Center for Computational Oncology (a collaboration between the Oden Institute for Computational Engineering and Sciences, The University of Texas MD Anderson Cancer Center, and Texas Advanced Computing Center).The project was also supported by the Tumor Measurement Initiative at The University of Texas MD Anderson Cancer Center.We acknowledge the support from “la Caixa†Foundation (ID 100010434), fellowship code is LCF/BQ/PI23/11970033.
竞争利益
Caroline Chung – Research funding to the institution from Siemens Healthineers and RaySearch Laboratories
附加信息
出版商的注释
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。权利和权限
开放访问
本文在Creative Commons Attribution-Noncormercial-Noderivatives 4.0国际许可下获得许可,该许可允许任何非商业用途,共享,分发和复制以任何媒介或格式的形式,只要您提供适当的原始作者和来源的信用,请符合原始作者和来源,并提供了与Creative Commons的链接,并指示您是否修改了许可的材料。您没有根据本许可证的许可来共享本文或部分内容的改编材料。The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.要查看此许可证的副本,请访问http://creativecommons.org/licenses/by-nc-nd/4.0/。重印和权限
引用本文
Wu, C., Andaloussi, M.A., Hormuth, D.A.
等。A critical assessment of artificial intelligence in magnetic resonance imaging of cancer.NPJ成像3 , 15 (2025).https://doi.org/10.1038/s44303-025-00076-0
已收到:
公认:
出版:
doi:https://doi.org/10.1038/s44303-025-00076-0