A call for built-in biosecurity safeguards for generative AI tools
作者:Church, George
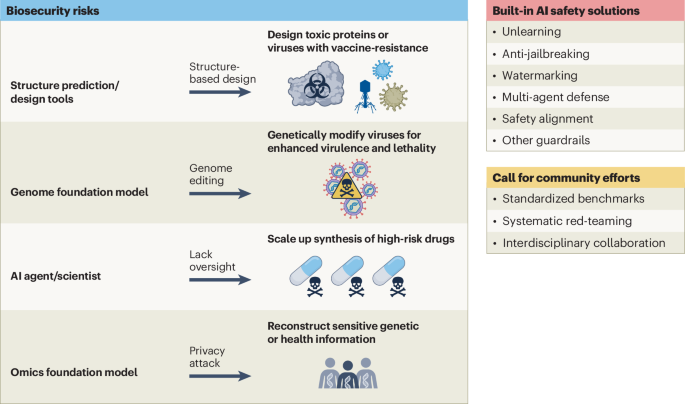
Generative AI is changing biotechnology research, and accelerating drug discovery, protein design and synthetic biology. It also enhances biomedical imaging, personalized medicine and laboratory automation, which enables faster and more efficient scientific advancements. However, these breakthroughs have also raised biosecurity concerns, which has prompted policy and community discussions1,2,3,4.
The power of generative AI lies in its ability to generalize from known data to the unknown. Deep generative models can predict novel biological molecules that might not resemble existing genome sequences or proteins. This capability introduces dual-use risks and serious biosecurity threats — such models could potentially bypass the established safety screening mechanisms used by nucleic acid synthesis providers5, which presently rely on database matching to identify sequences of concerns6. AI-driven tools could be misused to engineer pathogens, toxins or destabilizing biomolecules, and AI science agents could amplify risks by automating experimental designs7.
This is a preview of subscription content, access via your institution
References
Baker, D. & Church, G. Science 383, 349 (2024).
Bloomfield, D. et al. Science 385, 831–833 (2024).
Blau, W. et al. Proc. Natl Acad. Sci. USA 121, e2407886121 (2024).
Bengio, Y. et al. Preprint at https://doi.org/10.48550/arXiv.2501.17805 (2025).
Wittmann, B. J. et al. Preprint at bioRxiv https://doi.org/10.1101/2024.12.02.626439 (2024).
Fast Track Action Committee On Synthetic Nucleic Acid Procurement Screening. Framework For Nucleic Acid Synthesis Screening. (White House Office of Science and Technology Policy, 2024).
Boiko, D. A., MacKnight, R., Kline, B. & Gomes, G. Nature 624, 570–578 (2023).
Church, G. A synthetic biohazard non-proliferation proposal. Harvard Medical School https://arep.med.harvard.edu/SBP/Church_Biohazard04c.htm (2004).
Nguyen, E. et al. Science 386, eado9336 (2024).
Huang, K. et al. Preprint at https://doi.org/10.48550/arXiv.2404.18021 (2024).
Zhang, Z. et al. Preprint at https://doi.org/10.1101/2024.10.23.619960 (2024).
Rafailov, R. et al. Direct preference optimization: your language model is secretly a reward model. In NIPS’23: Proc. 37th International Conf. Neural Information Processing Systems (eds Oh, A.) 53728–53741 (Curran Associates, 2023).
Liu, S. et al. Nat. Mach. Intell. 7, 181–194 (2025).
Acknowledgements
Certain tools and software are identified in this Correspondence to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the tools and software identified are necessarily the best available for the purpose.
Ethics declarations
Competing interests
Z.Z., A.S.B., A.V., S.G., S.L.-G., S.C., M.B. and J.M. have no competing interests. E.X. has equity in GenBio AI. G.C. has biotechnology patents and equity in Lila.AI, DynoTx, Jura.bio, ShapeTx, GC-Tx, ArrivedAI, Nabla.bio, Manifold.bio and Plexresearch. M.W., L.C. and Y.Q. invented some of the technologies mentioned in this Correspondence, with patent applications filed by Princeton University and Stanford University. L.C. is scientific advisor to Acrobat Genomics and Arbor Biotechnologies.
About this article
Cite this article
Wang, M., Zhang, Z., Bedi, A.S. et al. A call for built-in biosecurity safeguards for generative AI tools. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02650-8
Published:
DOI: https://doi.org/10.1038/s41587-025-02650-8
