- 研究
- 开放访问
- 发布:
- 哈南·哈桑·艾哈迈德(Hanan Hassan Ahmed)2,,,,
- Radhwan Abdul Kareem3,,,,
- Anupam Yadav4,,,,
- Subbulakshmi Ganesan5,,,,
- 阿曼·尚克扬(Aman Shankhyan)6,,,,
- 索非亚·古普塔(Sofia Gupta)7,,,,
- Kamal Kant Joshi8,,,,9,,,,
- Hayder Naji Sameer10,,,,
- 艾哈迈德·亚森(Ahmed Yaseen)11,,,,
- Zainab H. Athab12,,,,
- Mohaned Adil13和
- …
- Bagher远一个 orcid:orcid.org/0000-0003-2290-722014一个
抽象的
客观的
这项研究旨在创建一个可靠的框架来对食管癌进行分级。该框架结合了特征提取,深度学习与注意机制和放射素学,以确保在肿瘤分析中的准确性,可解释性和实际使用。
材料和方法
这项回顾性研究使用了从2018年至2023年收集的多个临床中心的2,560例食管癌患者的数据。数据集包括CT扫描图像和临床信息,代表各种癌症等级和类型。遵循标准化的CT成像方案,并手动分割了经验丰富的放射科医生。研究中仅使用高质量数据。使用血清平台提取了总共215个放射线特征。该研究使用了两种深度学习模型的Densenet121和EfficityNet-B0增强了注意机制,以提高准确性。一种合并的分类方法使用了放射线和深度学习功能,并应用了随机森林,XGBOOST和CATBOOST等机器学习模型。通过严格的培训和测试程序对这些模型进行了验证,以确保有效的癌症分级。
结果
这项研究分析了对食管癌的放射素和深度学习特征的可靠性和性能。基于其ICC(类内相关)值将放射线特征分为四个可靠性水平。大多数功能具有出色的(ICC> 0.90)或良好(0.75 <icc的0.90)可靠性。还对从Densenet121和ExtricNet-B0提取的深度学习功能也进行了分类,其中一些表现出差的可靠性。包括XGBOOST和CATBOOST在内的机器学习模型,测试了其对癌症进行评分的能力。XGBoost具有递归特征消除(RFE)的XGBOOST为放射线特征提供了最佳效果,其AUC(曲线下方)为91.36%。对于深度学习功能,使用主组件分析(PCA)的XGBoost使用Densenet121获得了最佳结果,而使用RFE的Catboost则在ExtricNet-B0方面表现最佳,获得了94.20%的AUC。将放射线和深度特征结合起来导致了显着改善,XGBoost的AUC最高为96.70%,准确性为96.71%,灵敏度为95.44%。集合模型中的Densenet121和EfficityNet-B0模型的组合达到了最佳总体性能,AUC为95.14%,精度为94.88%。
结论
这项研究通过结合放射素学和深度学习来改善食道癌的分级。它提高了诊断准确性,可重复性和可解释性,同时还通过更好的肿瘤表征来帮助进行个性化治疗计划。
临床试验号
不适用。
介绍
食管癌仍然是肿瘤学的主要挑战,死亡率高和晚期诊断。准确的癌症分级对于制定个性化的治疗计划和改善患者预后至关重要[1,,,,2,,,,3,,,,4]。但是,传统的诊断方法通常会遭受主观解释和特征提取的不一致。为了克服这些问题,这项研究使用了先进的机器学习技术,包括放射素学和深度学习,以开发一个综合框架,以准确且可重复的食管癌分级[5,,,,6,,,,7]。
食管癌的早期和精确分级对于优化治疗计划和改善患者预后至关重要。高级肿瘤通常更具侵略性,具有更大的转移可能性,并且与生存率较差有关。通过在早期对肿瘤进行准确分类,临床医生可以制定个性化的治疗策略,以确保有效地使用手术,化学疗法和放射治疗来改善患者的预后。此外,早期分级有助于更好的风险评估和持续监测,从而及时进行干预,以防止疾病进展并增强长期生存。
食管癌在解剖学上分为肿瘤位置分为三个区域。食道上部从丝状软骨延伸到胸腔入口,食道中部位于胸腔入口和气管分叉之间,下食管从气管分叉延伸到胃食管的延伸[2,,,,8]。这些分裂在临床上具有重要意义,因为肿瘤位置会影响手术方法,淋巴结转移模式和整体治疗策略。例如,上食道上的肿瘤可能需要广泛的手术切除,而食道下食道的肿瘤通常涉及胃食管治疗。这项研究包括来自所有三个解剖区域的肿瘤,以确保全面评估,从而适用于各种临床病例。
放射素学提供了一种可靠的方法来从医学图像中提取定量特征,例如CT扫描[9,,,,10,,,,11,,,,12]。这些特征捕获了重要的肿瘤特征,例如形状,质地和强度,它们与临床结果密切相关[13,,,,14,,,,15,,,,16]。虽然放射线学在肿瘤学方面表现出希望,但它可能会受到手动分割的利益区域(ROI)的可变性的影响[ROIS)[17,,,,18,,,,19,,,,20]。为了解决这个问题,研究使用了多细分策略,在该策略中,每个肿瘤被分割了三次,以确保特征的可重复性和可靠性[21,,,,22,,,,23,,,,24]。这种方法降低了ROI描述的可变性,提高了放射线分析的鲁棒性和有效性[25]。
深度学习,尤其是卷积神经网络(CNN),通过使特征提取并实现端到端学习来彻底改变医学成像[14,,,,26,,,,27,,,,28,,,,29]。在这项研究中,诸如Densenet121和ExtricNet-B0之类的高级模型用于从成像数据中提取复杂的高维特征的能力。这些模型可有效捕获准确的食管癌分级所需的复杂模式。注意机制通过关注肿瘤内诊断重要的区域进一步增强模型,从而精确地分析微妙的纹理和形态学变化对分级至关重要[30,,,,31,,,,32]。与依赖于预定义特征的传统放射素学不同,深度学习直接从数据中学习了歧视性模式,从而消除了对手动特征选择的需求[11,,,,33]。此外,深度学习模型可以捕获低级细节,例如边缘和纹理,以及高级语义信息,例如肿瘤形态,从而可以完整地了解成像数据。注意机制通过优先考虑相关领域来提高可解释性和准确性。
虽然深度学习功能提供自适应,高维的见解,但放射线特征提供了临床上可解释的标记,这些标记通常与生物过程直接相关。这项研究结合了这两种利用其优势的方法。放射线特征提供可靠的,可解释的指标,而深度特征捕获复杂的非线性图案[8,,,,34,,,,35]。整合两者都可以提高诊断准确性,并更全面地了解肿瘤特征。
这项研究的重点是功能提取的可重复性。分割的变异性是医学成像中的一个常见问题,尤其是放射素学中。为了解决这个问题,使用一致的方案通过经过训练的注释者对每个肿瘤进行了三次分割,并从每个分割中提取特征[36,,,,37,,,,38]。这种多细分策略可确保放射线和深度学习特征的可重复性。仔细评估了重复分段的特征的一致性,以确认其可靠性和稳定性。
这项研究介绍了一个新的食管癌分级框架,该框架整合了放射线和深度学习特征,以解决现有诊断方法的局限性。该框架以两种方式使用深度学习模型:作为直接癌症分级的端到端管道,作为功能提取器,从中间层中提取深度特征,并在传统的机器学习分类器中使用以增强可解释性和性能。这种双重方法最大程度地提高了深度学习衍生的特征的潜力。这项研究的主要贡献包括:
-
1。
可再现的特征提取:多细分策略可确保放射线和深度学习特征的可靠性。通过分割肿瘤三次并从每种特征中提取特征,可最大程度地减少变异性,从而导致更加一致和可重现的结果。
-
2。
注意力增强的深度学习:densenet121和EfficityNet-B0模型,通过注意机制增强,重点是诊断至关重要的区域。这些机制通过优先考虑相关成像细节来提高模型的可解释性和准确性。
-
3。
功能集成:可提供临床上有用见解的放射线特征与高维的深度特征相结合。这种整合利用了两种方法的优势,从而产生了更强大,更全面的诊断框架。
-
4。
全面验证:该框架在食管癌病例的大型,多样化的数据集上进行了严格验证。评估了关键性能指标,例如准确性,灵敏度和可重复性,并与当前方法进行了比较,以确保临床可靠性和适用性。
材料和方法
研究设计和数据集
这项回顾性研究使用了来自2018年至2023年期间从多个临床中心收集的2,560名诊断为食管癌的患者的数据。数据集包括CT成像数据以及临床和病理信息,确保了患者群体的全面表示。癌症等级分布如下:384例(15%)为I级,896例患者(35%)为II级,640例患者(25%)为III级,640例患者(25%)(25%)为IV级。使用标准化的多探测器协议获取CT图像,该方案在中心之间具有一致的参数,例如切片厚度(3-5毫米),电压(120-140 kVp)和电流(100-300(100-300âmA))。对成像数据进行了预处理,以确保均匀性,包括分辨率归一化,对齐和去除由采集方案中的变化引起的伪影。所有患者数据均被匿名保护以保护机密性。随附的元数据包括人口统计细节,临床分期,肿瘤大小和组织学亚型。仔细策划数据集,以确保在癌症等级和肿瘤特征之间保持平衡的表示,从而为进一步的分析提供了坚实的基础。
包容和排除标准
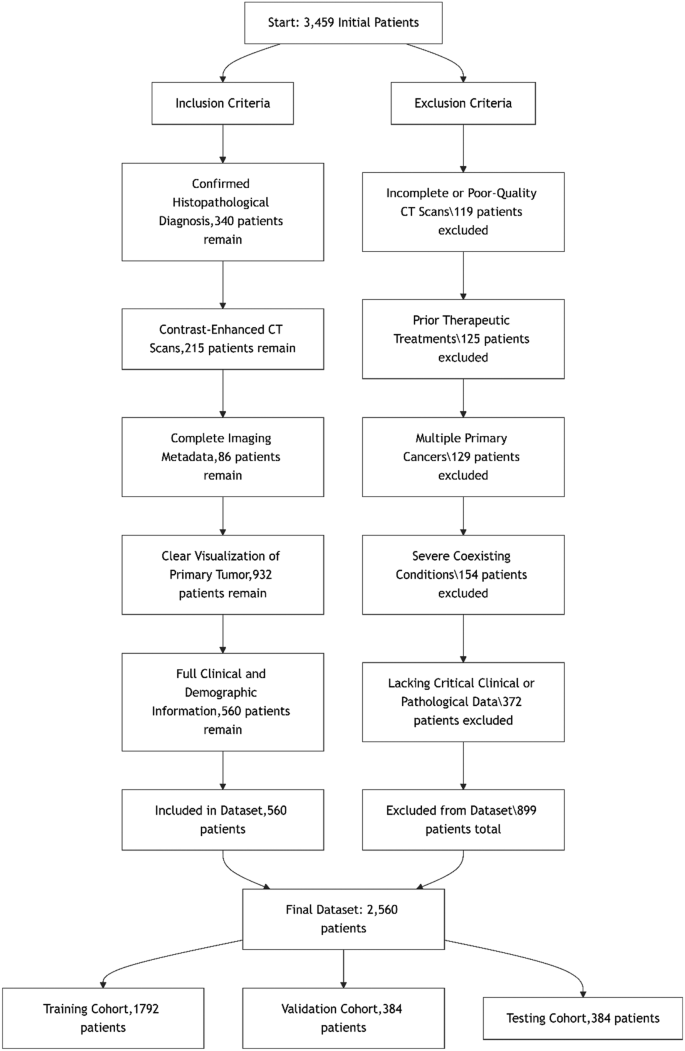
采用严格的包含和排除标准,以确保数据集的完整性和与食管癌分级的相关性。仅包括具有确认的组织病理学诊断,对比增强的CT扫描和完整临床数据的患者,以保持诊断准确性和一致性。排除标准消除了质量较差的案例,成像之前先前的肿瘤治疗,多种主要癌症或缺少关键数据,以确保高质量且可靠的数据集。补充材料中提供了这些标准的详细流程图。图 1说明了食管癌数据集的患者选择期间应用的严格纳入和排除标准。总共审查了3,459个初始患者记录。在应用严格的纳入和排除标准后,最终数据集包括2560名患者。纳入过程涉及选择确认的组织病理学诊断,对比增强的CT扫描,完整成像元数据,清除肿瘤可视化以及完整的临床和人口统计信息的患者。同时,由于不完整或质量较差的CT扫描,有899名患者被排除在外(n= 119),先前的治疗干预措施(n= 125),存在多种主要癌症(n= 129),严重合并症(n= 154),缺少关键的临床或病理数据(n= 372)。最终数据集随机分为三个同类:1,792例培训患者(70%),384名患者(15%)进行验证,384名患者(15%)进行测试,确保跨亚群的平衡级别分布并保留模型开发和评估的统计完整性。图1食管癌数据集的包含和排除标准为了更好地与二元分类任务保持一致并增强临床解释性,我们将患者分为低级(III级)和高级(III级IV级)类别。桌子 1
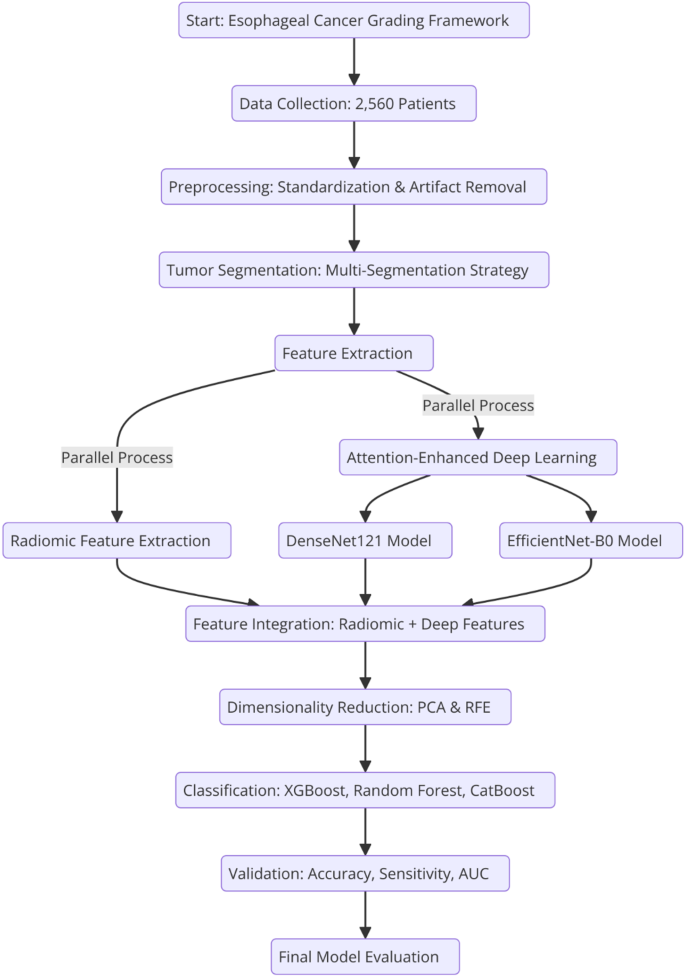
提出的框架结合了放射线和深度学习特征,以实现准确的食管癌分级。它遵循一种系统的方法,该方法始于肿瘤分割和放射素特征提取,然后将深度学习模型与注意机制的整合。该框架使用大型数据集经过全面的验证,以确保其临床适用性和癌症诊断中的可重复性。
2说明了从数据收集到最终模型评估的整个过程,强调了所涉及的关键步骤。图2
准确的肿瘤分割是图像预处理的关键步骤,直接影响放射线和深度学习工作流程特征提取的质量和可靠性。
在这项研究中,使用对比增强的CT扫描来描述与原发性肿瘤相对应的感兴趣区域(ROI)进行肿瘤分割。遵循手动分割协议,以确保整个成像数据集中的精确和标准化的描述。
为了提高鲁棒性和可重复性,使用了多细分策略。每个肿瘤都被经验丰富的放射科医生独立进行了三次,在肿瘤学成像方面至少具有五年的专业知识。这些分割间隔7天,以减少记忆偏见的可能性并确保独立评估。这种方法最大程度地减少了单个注释者引入的可变性和潜在偏见,从而达到了数据集的高度一致性。所有分割任务均使用3D Slicer(在医学成像中广受认可的工具)执行,以确保工作流程中的精度并保持均匀性。
分割后,采用后处理技术来标准化分段的ROI。这包括平滑肿瘤边界以消除不规则性,使像素强度归一化以解释CT采集设置的变化,并将分段切片跨不同成像平面对齐。这些程序确保了肿瘤ROI不仅准确,而且在整个数据集中也保持一致,从而适合下游放射线和深度学习分析。通过实施多细分策略和严格的后处理,本研究确保了肿瘤分割既可重复又可靠。这些高质量的分割为提取鲁棒成像功能,提高分析的有效性并确保数据集的整体鲁棒性奠定了基础。
放射素特征提取
使用标准化环境用于放射线分析(Sera)软件(Sera)软件(Sera)软件,从每个肿瘤的图像中提取了总共215个定量放射素特征(https://visera.ca/),遵循图像生物标准标准化计划(IBSI)的指南。这些功能包括79个一阶特征,捕获强度和形态特征,以及136个高阶3D特征,描述纹理和空间关系。特征提取遵循标准化的预处理步骤,以确保均匀的体素间距和强度归一化。为了优化模型性能,排除了差异较低或高相关性的功能。补充材料中提供了放射线特征类型的详细分解。
为了确保放射线和深度学习特征提取的一致性和准确性,将几个预处理步骤应用于CT图像。首先,使用高斯过滤器进行肿瘤边界平滑,以减少噪声,同时保留肿瘤的结构完整性。接下来,使用Z分数转换应用体素强度归一化,以在不同的扫描和成像中心进行标准化强度值。为了纠正采集设置的变化,进行了切片比对和重采样,以达到1毫米1毫米1 mm的均匀空间分辨率。此外,通过将半自动分割方法应用于掩盖非肿瘤区域并最大程度地减少背景噪声来进行伪影。这些预处理步骤确保了成像数据在患者之间进行协调,从而提高了功能可重复性和模型性能。
放射素特征的可重复性评估
使用精心选择的参数(包括双向随机效应模型,绝对一致性和多个评估者或测量值)评估了放射素特征的可重复性。ICC被广泛认为是用于评估连续变量之间一致性的鲁棒统计指数,尤其是在需要可重现测量的研究中。该指数范围从0到1,提供了可靠性的定量度量。
使用ICC评估了放射线特征的可靠性,遵循具有绝对一致的双向随机效应模型,以评估多个分段的一致性。特征是根据广泛接受的阈值进行分类的:良好的可靠性(ICC> 0.90),良好的可靠性(0.75 <icc -icc -0.90),中等可靠性(0.50 <icc -0.75)和可靠性差(ICC -0.50)。这些阈值是根据IBSI和以前的放射线学研究的准则选择的,以确保与现有文献的标准化和可比性。仅保留具有出色或良好可靠性的功能以进一步分析,以确保稳健和可重现的结果。使用为本研究开发的基于Python的内部代码进行ICC计算,以确保统计评估的精确性和一致性。该过程确保仅选择高度可靠的特征进行随后的建模和分析,从而增强了研究结果的鲁棒性。
深度学习框架
型号体系结构:Densenet121和Extricnet-B0
在这项研究中,使用了两种高级CNN架构,即Densenet121和EfficityNet-B0,用于分析CT成像数据。Densenet121采用密集连接的层,其中每层都以馈送方式直接连接到其他层。该体系结构启用功能重复使用,减少参数的数量并增强梯度流,从而使其对于具有有限数据集的医学成像任务特别有效。另一方面,有效网络-B0基于一种复合缩放方法,该方法均匀地缩放了网络深度,宽度和分辨率,并以优化的计算效率实现了最先进的精度。这些体系结构的互补优势选择:Densenet121的稳健特征传播和有效使用模型容量和可伸缩性。通过从ImageNet进行预训练的权重,以利用传递学习和加速收敛性初始化两种模型。
注意机制的实施
为了提高模型性能并专注于肿瘤内的重要区域,将注意力机制添加到了深度学习框架中。具体而言,使用了空间和通道注意模块。空间注意机制突出了CT图像中的关键区域,例如肿瘤的核心和边缘,这对于诊断很重要。通道注意机制通过对卷积层中最相关的通道增加重量来调整特征图,从而确保关键特征脱颖而出。通过整合这些注意力模块,可以提高食道癌分级的准确性,并且通过关注成像数据中肿瘤相关的区域,模型变得更加可解释。
深度提取
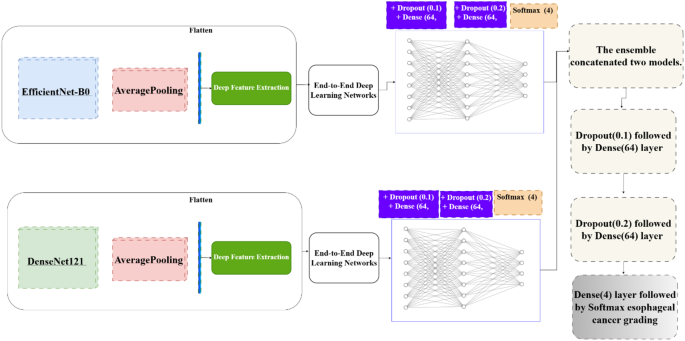
除了将Densenet121和EdgitionNet-B0用作端到端分类器外,还使用全球平均池提取了深度特征(图。 3)。然后将这些功能用作传统机器学习分类器的输入,并补充端到端方法。
深度提取过程遵循以下步骤:
-
1。
预处理CT图像并输入预先训练的Densenet121和有效网络B0模型。
-
2。
提取了中间层的特征图,从而捕获低级特征(例如边缘,纹理)和高级语义特征(例如肿瘤形状和空间关系)。
-
3。
这些特征被扁平化为特征向量。
-
4。
将功能向量馈入机器学习分类器,从而创建了一种混合方法,将深度学习与传统方法结合在一起。
这种双重方法端到端的深度学习和深度提取效果可确保灵活性,改善解释性和最大化的诊断性能。注意机制的添加进一步增强了这些模型,从而使系统能够专注于成像数据中最相关的部分。
功能集成和分类框架
为了提高诊断性能,提出的框架将放射线特征与深度学习衍生的特征相结合,结合了它们的互补优势。放射性特征以其临床解释性而闻名,捕获肿瘤的形态,强度和纹理,而从densenet121和ExcilityNet-B0模型中提取的深度特征捕获了复杂的层次结构和非线性模式。这种整合提供了肿瘤特征的全面表示,将特定于领域的知识与数据驱动的见解相结合。
鉴于集成特征集的高维度,降低维度降低和特征选择技术用于提高计算效率并降低过度拟合的风险。使用递归特征消除(RFE)和主成分分析(PCA)进行降低降低。RFE被用来迭代地删除非限制功能,同时保持预测精度。PCA将高维的放射线和深度学习特征转化为正交组件,从而捕获数据集中的最大差异。选择这些方法是因为它们在缓解过度拟合的同时提高分类性能的能力。尽管存在诸如LASSO回归和基于信息的选择之类的替代方法,但RFE和PCA为这项研究的数据集特性提供了最佳的特征选择。
分类框架采用了三种高级机器学习模型:随机森林,XGBOOST和CATBOOST。随机森林是一种基于决策树的合奏学习方法,其能够通过平均多个基于基于树的模型来提供强大的预测和抵抗过度适应的能力而闻名。XGBOOST是一种提高梯度的决策树算法,高效,通过在训练过程中优化速度和准确性,在结构化数据上表现出色。Catboost是另一种梯度增强算法,旨在有效地处理分类功能,即使在复杂的数据集中,也可以最大程度地减少过度拟合,同时可提供高精度。选择了这些模型的互补优势,确保了可靠的,高性能的分类管道,用于食管癌分级。
培训和验证管道
集成特征集分为三个子集:训练70%,验证15%,测试15%。该部门确保对分类模型进行彻底评估。对训练组进行了培训,并使用网格搜索和交叉验证对其超参数进行了微调,以获得最佳性能。验证子集用于基于关键指标(例如精度,灵敏度以及接收器操作特征曲线(AUC))的区域来评估模型。这种方法有助于确保模型在新的,看不见的数据上表现良好,同时仍提供准确的诊断结果。通过将放射线和深度学习功能结合起来,以及用于降低数据复杂性并使用功能强大的机器学习模型的先进技术,提出的框架为食管癌分级提供了全面,可扩展且易于理解的解决方案。
高参数调整和优化
为了提高机器学习和深度学习模型的性能,进行了高参数调整。对于XGBoost,Catboost和Random Forest等机器学习模型,使用5倍交叉验证的网格搜索来找到重要参数的最佳设置,例如估计器数量,学习率和最大深度。对于诸如Densenet121和ExtricNet-B0之类的深度学习模型,使用网格搜索和贝叶斯优化的组合对批量大小,学习率,辍学率以及完全连接的层的数量进行调整。
选择ADAM优化器的初始学习率为1E-4。还使用了早期停止机制来监视验证损失并防止过度拟合。在单独的测试集中评估了所有模型,最多可用于1000个时期,并在单独的测试集中评估关键性能指标,例如准确性,灵敏度和AUC,以确保这些模型在新数据上的性能良好。这些调整技术在确保研究结果强大,有效且可重现方面起着至关重要的作用。
硬件和软件规格
实验是在包括NVIDIA TESLA V100 GPU(32 GB VRAM),双Intel Xeon Silver 4210处理器和256 GB RAM的高性能计算系统上进行的。该系统运行Ubuntu 20.04,确保它与最新版本的深度学习框架兼容。所有模型均使用Python 3.9以及Tensorflow 2.8,Pytorch 1.12和Scikit-Learn 1.1等文库开发。使用3D切片机和放射线分析(Sera)工具包进行图像预处理和放射线特征提取。CUDA 11.6和CUDNN 8.3的使用有助于优化GPU加速,以实现深度学习任务。
结果
基于ICC值的可靠性级别
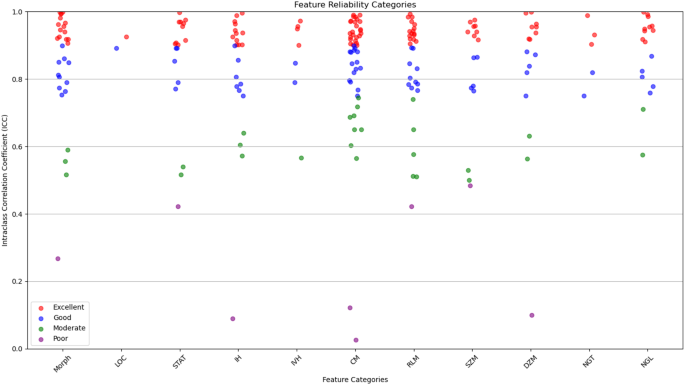
在年龄,性别,肿瘤大小和组织学亚型的低度和高级组之间观察到统计学上的显着差异(p<<0.05),高级肿瘤与年龄较大,男性,大小和腺癌组织学更为频繁。No significant difference was found in tumor location distribution.This stratification allows for a more clinically meaningful evaluation of model performance and reflects the grading criteria utilized during model development.Statistical analysis of the clinical and demographic characteristics revealed significant differences in age, tumor size, and histological subtype across esophageal cancer grades.Patients with higher-grade tumors tended to be older (p < 0.001) and had significantly larger tumor sizes (p < 0.001), indicating a potential association between tumor progression and increased patient age.Additionally, the prevalence of adenocarcinoma increased in more advanced stages (p = 0.003), suggesting histological shifts as the disease progresses.However, no significant differences were observed in gender distribution (p = 0.12) or tumor location (p = 0.09), implying that these factors do not strongly influence cancer grade.These findings provide insights into the clinical progression of esophageal cancer and highlight the importance of tumor size and histology in disease severity assessment.The radiomic features were categorized based on their reliability, as determined by their ICC values.It breaks down the distribution of features, indicating how many had excellent, good, moderate, or poor reliability.
This categorization is crucial for ensuring that only the most reliable features are used in further analyses, which strengthens the overall reliability of the study.Out of the 215 features evaluated, most were found to have excellent reliability (ICC > 0.90) or good reliability (0.75 < ICC ≤ 0.90).These reliable features were spread across 11 categories, including tumor morphology, local intensity, and various texture metrics like co-occurrence matrix and size zone matrix features.To evaluate inter-observer variability, tumor segmentation was performed independently by three experienced radiologists at different time points.The agreement between segmentations was quantified using the Dice Similarity Coefficient (DSC) and ICC.The obtained DSC values (≥ 0.85) and ICC values (> 0.90) confirmed high consistency in the multi-segmentation strategy, ensuring that feature extraction was reliable and reproducible across different annotators.
A correlation analysis was conducted to examine the relationship between radiomic features and key clinical variables, including tumor grade, tumor size, patient age, and tumor location.Spearman’s correlation coefficient was used to assess statistical associations.The analysis revealed that tumor size had a strong positive correlation with first-order intensity features (Ï = 0.62,p < 0.001), suggesting that larger tumors exhibit distinct intensity distributions.Tumor grade showed a significant correlation with texture-based radiomic features, particularly co-occurrence matrix features (Ï = 0.54,p < 0.01), indicating that higher-grade tumors tend to have greater heterogeneity.A weak but statistically significant correlation was observed between patient age and morphology features (Ï = 0.21,p = 0.03), suggesting subtle structural differences in tumors among older patients.However, no significant correlation was found between tumor location and radiomic feature distribution (p > 0.05), implying that textural and morphological characteristics are relatively consistent across different anatomical regions of the esophagus.These findings highlight the potential of radiomic features as imaging biomarkers for tumor characterization and further support their clinical relevance in esophageal cancer grading.A detailed summary of correlation coefficients and p-values is provided in the supplementary materials.
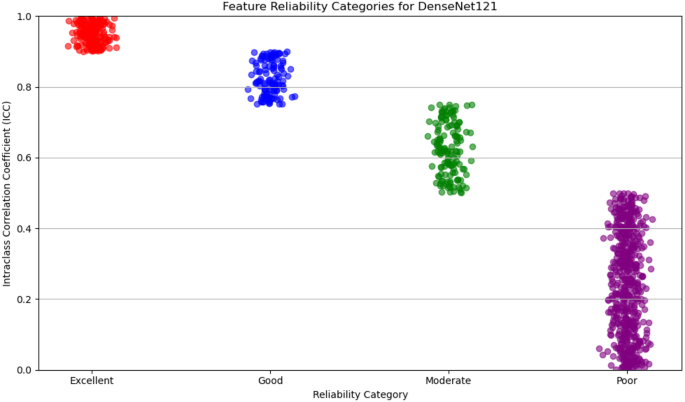
数字4illustrates the distribution of radiomic features across the four reliability categories (excellent, good, moderate, and poor) based on ICC values.The plot provides a visual representation of the robustness of features within each category, emphasizing the predominance of features with high reliability.This visualization underscores the careful selection of reproducible features, ensuring the robustness of the study’s analytical framework.
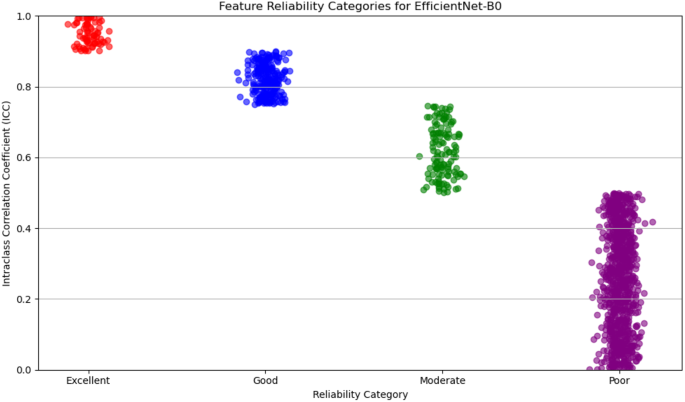
For deep feature extraction, features were obtained from the Global Average Pooling layers of two pre-trained networks: DenseNet121 and EfficientNet-B0.DenseNet121 produced 1,024 features from its Global Average Pooling layer, while EfficientNet-B0 generated 1,280 features.These features capture complex, high-dimensional representations of tumor characteristics and were then categorized based on their reliability, which was assessed in terms of discriminative power and reproducibility.The reliability of features from both DenseNet121 and EfficientNet-B0 was evaluated and categorized into four levels: excellent, good, moderate, and poor.
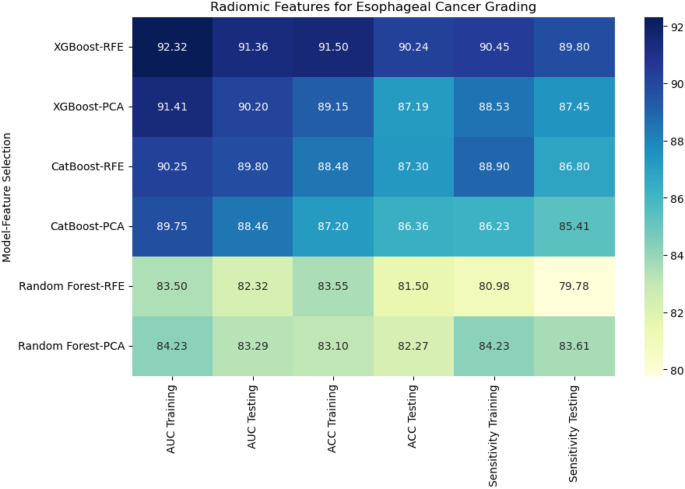
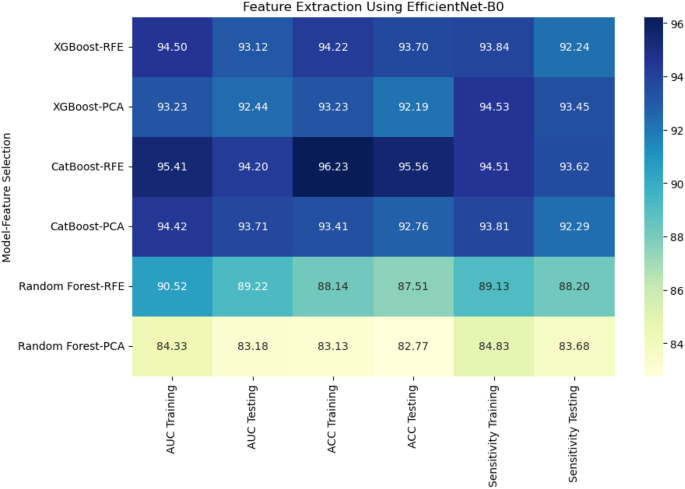
For DenseNet121, 53% of the extracted features were classified as poor reliability, while 21% were considered excellent, 12% good, and 14% moderate.On the other hand, EfficientNet-B0 showed a higher proportion of features with poor reliability, with 65% falling into this category.The remaining features were distributed as 6% excellent, 19% good, and 10% moderate.Figures 5和6display the reliability distribution of the deep features extracted from DenseNet121 and EfficientNet-B0, respectively.Esophageal cancer grading: model performance comparisonFor radiomic features, XGBoost with Recursive Feature Elimination (RFE) showed the best performance, achieving an AUC of 91.36%, an accuracy of 90.24%, and a sensitivity of 89.80% on the testing set (Fig. 7
)。
CatBoost combined with Principal Component Analysis (PCA) performed slightly lower, with an AUC of 88.46% and accuracy of 86.36%.Random Forest, even with PCA feature selection, was the weakest performer, with an AUC of 83.29% and accuracy of 82.27%.This result highlights the limitations of Random Forest when handling high-dimensional radiomic features.
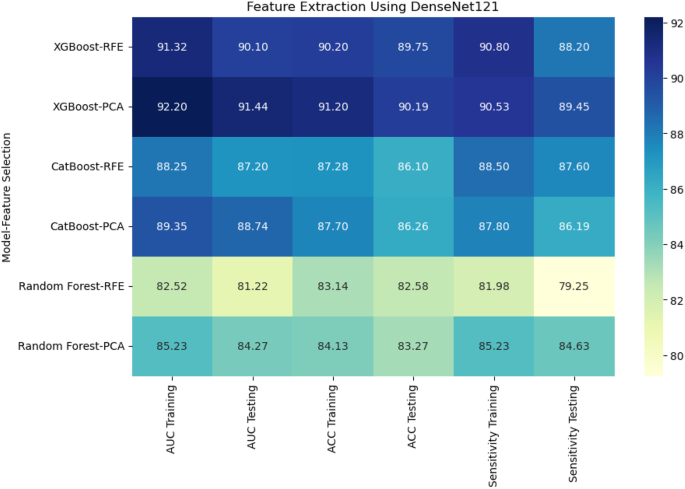
When deep features were extracted using DenseNet121, XGBoost with PCA again performed the best, achieving an AUC of 91.44% and accuracy of 90.19% on the testing set.CatBoost with RFE achieved an AUC of 87.20% and accuracy of 86.10%, while Random Forest with PCA had a lower performance with an AUC of 84.27% and accuracy of 83.27% (Fig. 8)。Using EfficientNet-B0 for deep feature extraction, similar trends were observed.CatBoost with RFE outperformed the other models, reaching an AUC of 94.20% and accuracy of 95.56% (Fig. 9)。XGBoost with PCA followed closely, achieving an AUC of 92.44% and accuracy of 92.19%.Random Forest continued to underperform with an AUC of 83.18% and accuracy of 82.77%.
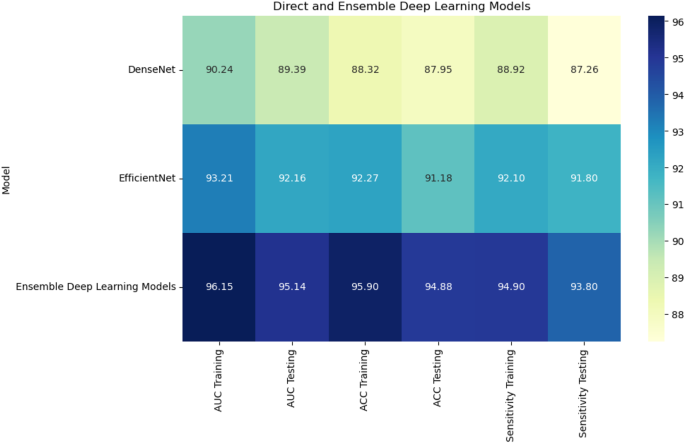
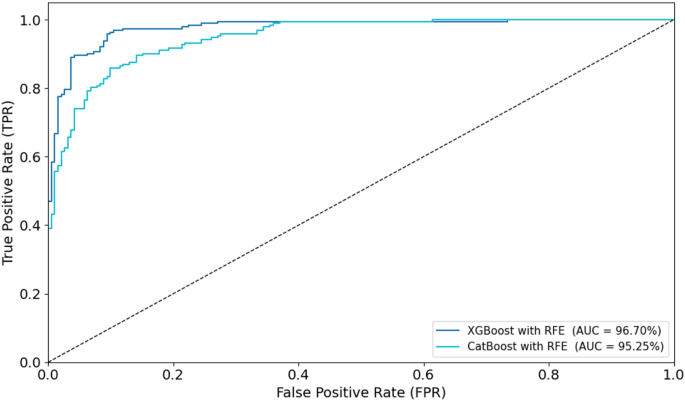
Combining radiomic and deep features resulted in significant improvements across models.XGBoost with RFE on combined features achieved the highest AUC of 96.70%, accuracy of 96.71%, and sensitivity of 95.44% on the testing set (Fig. 10)。CatBoost with RFE also performed well, with an AUC of 95.25% and accuracy of 95.26%.Random Forest with PCA again lagged behind, achieving an AUC of 87.28% and accuracy of 88.17%.In a comparison of direct deep learning models, EfficientNet-B0 outperformed DenseNet121, achieving an AUC of 92.16% and accuracy of 91.18% (Fig. 11)。However, ensemble models combining both DenseNet121 and EfficientNet-B0 features provided the best performance, with an AUC of 95.14%, accuracy of 94.88%, and sensitivity of 93.80% on the testing set.This demonstrates the superior ability of ensemble models to enhance predictive accuracy in esophageal cancer grading.
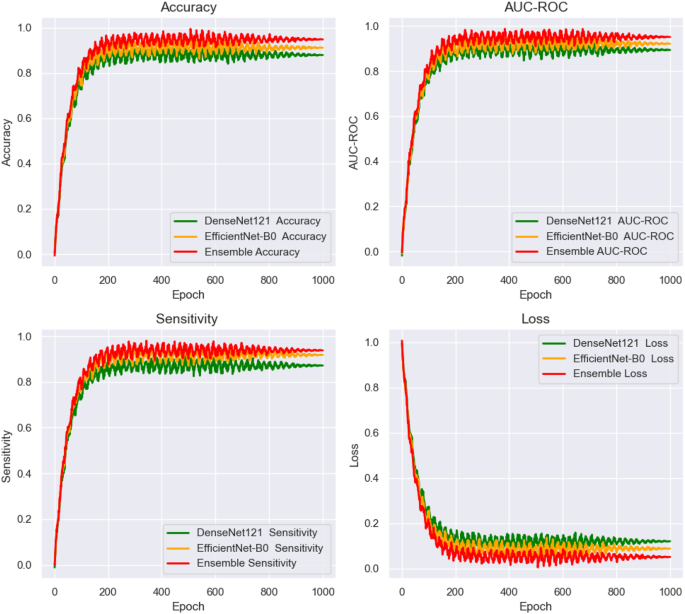
数字12shows the training and loss curves for the end-to-end deep learning networks, plotted over 1000 epochs.The curves track the performance of the networks across three key evaluation metrics: AUC, accuracy, and sensitivity.These metrics were closely monitored during the training process, providing valuable insights into the model’s convergence behavior and overall stability.The training curves illustrate the gradual improvement in performance over time, highlighting the model’s ability to learn from the data.The loss curves demonstrate the reduction in error throughout the training phase, indicating how well the models were optimizing.This figure enables a direct comparison of the models’ learning capabilities and generalization.It also shows which deep learning models exhibited the most efficient convergence and consistent performance across the full 1000 epochs, helping to assess their robustness and ability to handle the task.
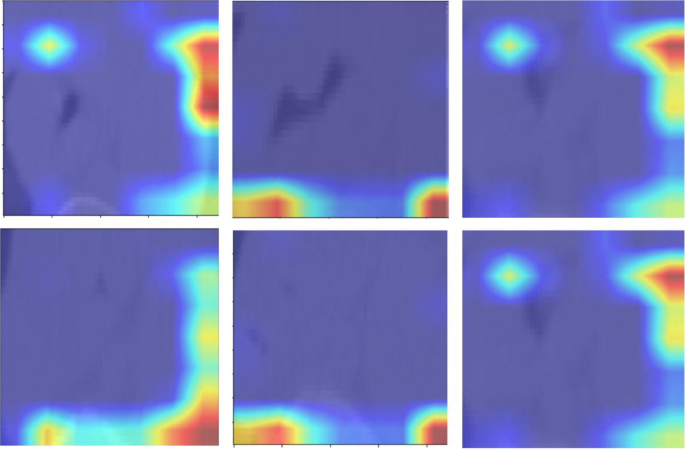
数字13presents the attention maps generated by the transformer-based models, overlaid on the corresponding images.These maps highlight the regions of interest that the models focus on during the classification process.By incorporating attention mechanisms, the models were able to emphasize critical areas of the image that are essential for accurate tumor grading in esophageal cancer.The attention maps visually illustrate how the models prioritize specific features, offering a more interpretable view of the deep learning decision-making process.These maps reveal which anatomical structures the model deems most relevant for making accurate predictions.By providing insights into the model’s focus, the attention maps help to enhance the interpretability of the deep learning models, contributing to their overall accuracy and robustness in classifying esophageal cancer.It is important to note that the attention maps presented in Fig. 13reflect cropped image regions centered around the segmented tumors.The areas of focus, while appearing to lie outside the tumor in some cases, are in fact within the peritumoral zone included in the segmentation masks.These regions were retained due to their diagnostic value in capturing surrounding textural variations.The attention mechanisms accurately highlight these intratumoral and peritumoral features, reinforcing the model’s interpretability and its ability to localize clinically relevant structures for esophageal cancer grading.
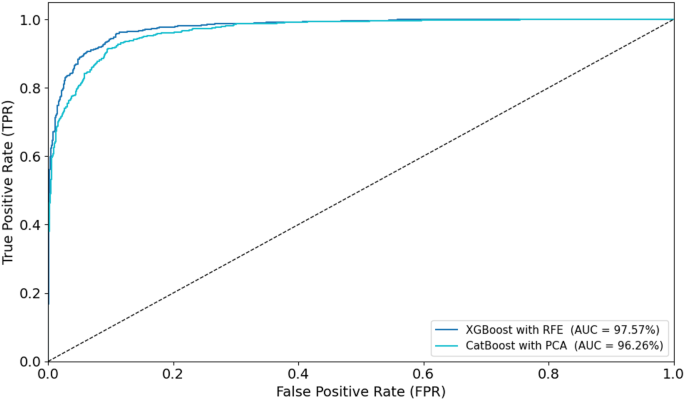
To visually assess classification performance, ROC curves were plotted for the optimal models during training and testing (Figs. 14和15)。In the training phase, the XGBoost with RFE model achieved the highest AUC of 97.57%, followed closely by CatBoost with PCA (AUC = 96.26%).For the testing set, XGBoost with RFE again outperformed other models with an AUC of 96.70%, while CatBoost with RFE achieved an AUC of 95.25%.These ROC curves, displayed in Figs. 14和15, provide visual confirmation of the strong discriminatory ability of these models in distinguishing between low- and high-grade esophageal cancers.
讨论
This study presents a novel approach for esophageal cancer grading by integrating radiomic features with deep learning techniques, enhanced by attention mechanisms.The goal is to improve diagnostic accuracy and reproducibility, addressing the limitations of traditional diagnostic methods, which often suffer from subjective interpretation and inconsistent feature extraction.Our framework leverages both radiomic features, which are interpretable and clinically relevant, and deep learning models—specifically DenseNet121 and EfficientNet-B0—that capture complex tumor characteristics directly from imaging data.The attention mechanism refines this process by focusing the model’s attention on diagnostically significant regions, enhancing prediction accuracy and ensuring that subtle morphological and textural variations are properly analyzed.Additionally, the multi-segmentation strategy employed in this study improves the reproducibility of the extracted features, ensuring the robustness of results and reducing variability, which is a common challenge in medical image analysis.To ensure optimal deep feature extraction, this study utilized DenseNet121 and EfficientNet-B0, two well-established architectures in medical imaging.DenseNet121’s densely connected layers improve gradient flow and enhance feature reuse, making it highly efficient for learning complex imaging patterns.EfficientNet-B0, leveraging a compound scaling approach, achieves high classification accuracy with reduced computational cost.These models were selected due to their complementary strengths in feature extraction and classification.The results demonstrated that integrating both networks in an ensemble framework further improved cancer grading performance.Future work may explore additional architectures, such as ResNet and Inception, to assess generalizability across different datasets.
When compared to existing studies in the field, our approach demonstrates notable improvements in several areas, including model performance, interpretability, and clinical applicability.For model performance, our study shows a significant enhancement in accuracy and reproducibility compared to previous works in radiomics and deep learning for esophageal cancer prediction.例如,Xie等。[39]combined radiomics and deep learning for predicting radiation esophagitis in esophageal cancer patients treated with volumetric modulated arc therapy (VMAT), achieving an AUC of 0.805 in external validation.While promising, their model was limited by dose-based features and lacked attention mechanisms to focus on relevant regions.Our approach integrates attention-enhanced deep learning, improving diagnostic accuracy and model interpretability, allowing clinicians to better understand the decision-making process.
Furthermore, our framework’s multi-segmentation strategy strengthens the model’s generalizability and minimizes variability arising from image segmentation inconsistencies.This approach is superior to other models, such as the one developed by Chen et al.[40] for predicting lymph node metastasis in esophageal squamous cell carcinoma (ESCC), which combined handcrafted radiomic features with deep learning but did not control for segmentation variability.Our multi-segmentation ensures that features are both reliable and robust across repeated segmentations, a crucial aspect for clinical applications where precision is key.
A significant strength of our study is the integration of radiomic and deep learning features.This combined approach has demonstrated superior performance in various oncology-related studies.例如,杨等人。[41] showed that CT-based radiomics could predict the T stage and tumor length in ESCC with AUC values of 0.86 and 0.95, respectively.However, their reliance on handcrafted radiomic features limited the ability to capture complex, non-linear tumor characteristics.Our framework, in contrast, combines the interpretability of radiomics with the high-dimensional, hierarchical feature extraction capabilities of deep learning models.This integration allows for more accurate predictions of cancer grading and provides a comprehensive representation of tumor characteristics, which is essential for personalizing treatment strategies.The combination of deep learning and radiomics is particularly important in clinical decision-making.Radiomic features, such as texture and shape, provide clinicians with meaningful insights into tumor biology.Meanwhile, the deep features extracted from DenseNet121 and EfficientNet-B0 capture subtle, complex patterns that are not easily identified through traditional methods.This dual approach offers a more comprehensive view of the tumor, supporting more informed and effective clinical decisions.
Reproducibility is another key advantage of our study.Variability in tumor segmentation is a significant issue in medical imaging, as it can introduce errors that compromise the reliability of predictive models.Several studies, such as those by Li et al.[40] and Du et al.[42], have developed radiomic models for esophageal cancer diagnosis, but none have applied as rigorous a multi-segmentation strategy as our study.We mitigate segmentation variability by performing three separate segmentations for each tumor and evaluating the consistency of the extracted features.This ensures that the data fed into both radiomics and deep learning models is reliable, which enhances the reproducibility of the final predictions.
Additionally, our study features comprehensive validation across a large and diverse dataset, ensuring robust performance across various patient populations.This stands in contrast to other studies, like Chen et al.[40], which primarily evaluated model performance within a single institution.Our validation process not only benchmarks performance in terms of accuracy, sensitivity, and specificity but also thoroughly assesses the model’s clinical applicability.In summary, the combination of radiomic and deep learning features, the integration of attention mechanisms, and the multi-segmentation strategy make our approach highly effective for esophageal cancer grading.It addresses the challenges faced by traditional methods and enhances the clinical applicability, interpretability, and reproducibility of esophageal cancer prediction.
Despite the promising results of our proposed framework, several limitations need to be considered.First, the framework depends on high-quality annotated datasets for both radiomic feature extraction and deep learning model training.Obtaining such datasets can be difficult, and variations in tumor segmentation and image quality across different clinical settings may affect the consistency of the results.Moreover, the model’s ability to generalize could be limited by the demographic and clinical diversity of the dataset used.To improve this, future studies should aim to test the model across multiple centers and more diverse patient populations to ensure its effectiveness and applicability in real-world clinical environments.
Another area for improvement is the inclusion of additional imaging modalities, such as PET or MRI scans, which could provide extra valuable information and further improve the model’s diagnostic accuracy.Future research could explore how these modalities could be integrated into the framework.Additionally, using more advanced attention mechanisms could improve the model’s interpretability and decision-making processes.This would allow healthcare providers to better understand why the model makes certain predictions, which is key for clinical adoption.Lastly, long-term validation of the framework’s impact on treatment outcomes and patient prognosis is essential.It is also important to investigate the possibility of real-time implementation of the model in clinical workflows to evaluate its practical use in supporting clinical decision-making.
结论
In conclusion, our study marks a significant step forward in esophageal cancer grading.By combining the strengths of radiomics and deep learning, we have been able to achieve higher diagnostic accuracy, reproducibility, and interpretability.The integration of attention mechanisms, the use of a multi-segmentation strategy, and the combination of radiomic features with deep learning insights have allowed us to develop a framework that outperforms previous models, predicting tumor characteristics with greater precision.This approach not only improves our understanding of tumor behavior but also holds great potential for optimizing personalized treatment plans for patients with esophageal cancer.
数据可用性
在当前研究中使用和/或分析的数据集可根据合理的要求从通讯作者获得。
参考
DiSiena M, Perelman A, Birk J, Rezaizadeh H. Esophageal cancer: an updated review.South Med J. 2021;114(3):161–8.
文章一个 PubMed一个 Google Scholar一个
Alsop BR, Sharma P. Esophageal cancer.Gastroenterol Clin.2016;45(3):399–412.
文章一个 Google Scholar一个
Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors.Clin J Gastroenterol.2020;13(6):1010–21.
文章一个 PubMed一个 Google Scholar一个
Li J, Xu J, Zheng Y, Gao Y, He S, Li H, et al.Esophageal cancer: epidemiology, risk factors and screening.Chin J Cancer Res.2021;33(5):535.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Xie CY, Pang CL, Chan B, Wong EYY, Dou Q, Vardhanabhuti V. Machine learning and radiomics applications in esophageal cancers using non-invasive imaging methods—a critical review of literature.癌症(巴塞尔)。2021;13(10):2469.
文章一个 PubMed一个 Google Scholar一个
Gong X, Zheng B, Xu G, Chen H, Chen C. Application of machine learning approaches to predict the 5-year survival status of patients with esophageal cancer.J Thorac Dis。2021;13(11):6240.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Hosseini F, Asadi F, Emami H, Ebnali M. Machine learning applications for early detection of esophageal cancer: a systematic review.BMC Med Inf Decis Mak.2023;23(1):124.
文章一个 Google Scholar一个
Wang Y, Yang W, Wang Q, Zhou Y. Mechanisms of esophageal cancer metastasis and treatment progress.前免疫。2023;14:1206504.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Salmanpour M, Hosseinzadeh M, Rezaeijo S, Ramezani M, Marandi S, Einy M et al.Deep versus handcrafted tensor radiomics features: Application to survival prediction in head and neck cancer.In: European Journal Of Nuclear Medicine And Molecular Imaging.Springer One New York Plaza, Suite 4600, New York, NY, United States;2022. pp. S245–6.
Salmanpour MR, Rezaeijo SM, Hosseinzadeh M, Rahmim A. Deep versus handcrafted tensor radiomics features: prediction of survival in head and neck Cancer using machine learning and fusion techniques.诊断。2023;13(10):1696.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Salmanpour MR, Hosseinzadeh M, Akbari A, Borazjani K, Mojallal K, Askari D, et al.Prediction of TNM stage in head and neck cancer using hybrid machine learning systems and radiomics features.Medical imaging 2022: Computer-Aided diagnosis.SPIE;2022. pp. 648–53.
Fatan M, Hosseinzadeh M, Askari D, Sheikhi H, Rezaeijo SM, Salmanpour MR.In: Andrearczyk V, Oreiller V, Hatt M, Depeursinge A, editors.Segmentation and Outcome Prediction.CHAM:Springer International Publishing;2022. pp. 211–23.Fusion-Based Head and Neck Tumor Segmentation and Survival Prediction Using Robust Deep Learning Techniques and Advanced Hybrid Machine Learning Systems BT - Head and Neck Tumor.
Cui Y, Li Z, Xiang M, Han D, Yin Y, Ma C. Machine learning models predict overall survival and progression free survival of non-surgical esophageal cancer patients with chemoradiotherapy based on CT image radiomics signatures.Radiat Oncol.2022;17(1):212.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wang J, Zeng J, Li H, Yu X. A deep learning radiomics analysis for survival prediction in esophageal cancer.J Healthc Eng。2022;2022(1):4034404.
Kawahara D, Murakami Y, Tani S, Nagata Y. A prediction model for degree of differentiation for resectable locally advanced esophageal squamous cell carcinoma based on CT images using radiomics and machine-learning.Br J Radiol.2021;94(1124):20210525.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Li L, Qin Z, Bo J, Hu J, Zhang Y, Qian L, et al.Machine learning-based radiomics prognostic model for patients with proximal esophageal cancer after definitive chemoradiotherapy.Insights Imaging.2024;15:284.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
AkbarnezhadSany E, EntezariZarch H, AlipoorKermani M, Shahin B, Cheki M, Karami A, et al.YOLOv8 outperforms traditional CNN models in mammography classification: insights from a Multi-Institutional dataset.Int J Imaging Syst Technol.2025;35(1):e70008.
文章一个 Google Scholar一个
Javanmardi A, Hosseinzadeh M, Hajianfar G, Nabizadeh AH, Rezaeijo SM, Rahmim A, et al.Multi-modality fusion coupled with deep learning for improved outcome prediction in head and neck cancer.Medical imaging 2022: image processing.SPIE;2022. pp. 664–8.
Rezaeijo SM, Harimi A, Salmanpour MR.Fusion-based automated segmentation in head and neck cancer via advance deep learning techniques.3D head and neck tumor segmentation in PET/CT challenge.Springer;2022. pp. 70–6.
Bijari S, Rezaeijo SM, Sayfollahi S, Rahimnezhad A, Heydarheydari S. Development and validation of a robust MRI-based nomogram incorporating radiomics and deep features for preoperative glioma grading: a multi-center study.定量成像Med Surg。2025;15(2):1121138–5138.
文章一个 Google Scholar一个
Xue C, Yuan J, Lo GG, Chang ATY, Poon DMC, Wong OL, et al.Radiomics feature reliability assessed by intraclass correlation coefficient: a systematic review.定量成像Med Surg。2021;11(10):4431.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Akinci D’Antonoli T, Cavallo AU, Vernuccio F, Stanzione A, Klontzas ME, Cannella R, et al.Reproducibility of radiomics quality score: an intra-and inter-rater reliability study.EUR RADIOL。2024;34(4):2791–804.
文章一个 PubMed一个 Google Scholar一个
Gong J, Wang Q, Li J, Yang Z, Zhang J, Teng X, et al.Using high-repeatable radiomic features improves the cross-institutional generalization of prognostic model in esophageal squamous cell cancer receiving definitive chemoradiotherapy.Insights Imaging.2024;15(1):239.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ma D, Zhou T, Chen J, Chen J. Radiomics diagnostic performance for predicting lymph node metastasis in esophageal cancer: a systematic review and meta-analysis.BMC Med成像。2024;24(1):144.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Bijari S, Sayfollahi S, Mardokh-Rouhani S, Bijari S, Moradian S, Zahiri Z, et al.Radiomics and deep features: robust classification of brain hemorrhages and reproducibility analysis using a 3D autoencoder neural network.Bioengineering.2024;11(7):643.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sui H, Ma R, Liu L, Gao Y, Zhang W, Mo Z. Detection of incidental esophageal cancers on chest CT by deep learning.前Oncol。2021;11:700210.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Islam MM, Poly TN, Walther BA, Yeh CY, Seyed-Abdul S, Li YC, et al.Deep learning for the diagnosis of esophageal cancer in endoscopic images: a systematic review and meta-analysis.癌症(巴塞尔)。2022;14(23):5996.
文章一个 PubMed一个 Google Scholar一个
Takeuchi M, Seto T, Hashimoto M, Ichihara N, Morimoto Y, Kawakubo H, et al.Performance of a deep learning-based identification system for esophageal cancer from CT images.食管。2021;18:612–20.
文章一个 PubMed一个 Google Scholar一个
Huang C, Dai Y, Chen Q, Chen H, Lin Y, Wu J, et al.Development and validation of a deep learning model to predict survival of patients with esophageal cancer.前Oncol。2022;12:971190.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Chen C, Ma Y, Zhu M, Yan Z, Lv X, Chen C, et al.A new method for Raman spectral analysis: decision fusion-based transfer learning model.J Raman Spectrosc.2023;54(3):314–23.
文章一个 CAS一个 Google Scholar一个
Rezvy S, Zebin T, Braden B, Pang W, Taylor S, Gao XW.Transfer learning for Endoscopy disease detection and segmentation with mask-RCNN benchmark architecture.In: CEUR Workshop Proceedings.CEUR-WS;2020. pp. 68–72.
Ling Q, Liu X. Image recognition of esophageal Cancer based on ResNet and transfer learning.Int Core J Eng.2022;8(5):863–9.
Mahboubisarighieh A, Shahverdi H, Jafarpoor Nesheli S, Alipoor Kermani M, Niknam M, Torkashvand M, et al.Assessing the efficacy of 3D Dual-CycleGAN model for multi-contrast MRI synthesis.Egypt J Radiol Nucl Med.2024;55(1):1–12.
文章一个 Google Scholar一个
Huang G, Zhu J, Li J, Wang Z, Cheng L, Liu L, et al.Channel-attention U-Net: channel attention mechanism for semantic segmentation of esophagus and esophageal cancer.IEEE访问。2020;8:122798–810.
文章一个 Google Scholar一个
Cai Y, Wang Y. Ma-unet: An improved version of unet based on multi-scale and attention mechanism for medical image segmentation.In: Third international conference on electronics and communication;network and computer technology (ECNCT 2021).SPIE;2022. pp. 205–11.
Mali SA, Ibrahim A, Woodruff HC, Andrearczyk V, Müller H, Primakov S, et al.Making radiomics more reproducible across scanner and imaging protocol variations: a review of harmonization methods.J Pers Med。2021;11(9):842.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Pfaehler E, Zhovannik I, Wei L, Boellaard R, Dekker A, Monshouwer R, et al.A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features.Phys Imaging Radiat Oncol.2021;20:69–75.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Jha AK, Mithun S, Jaiswar V, Sherkhane UB, Purandare NC, Prabhash K, et al.Repeatability and reproducibility study of radiomic features on a Phantom and human cohort.Sci Rep. 2021;11(1):2055.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Xie C, Yu X, Tan N, Zhang J, Su W, Ni W, et al.Combined deep learning and radiomics in pretreatment radiation esophagitis prediction for patients with esophageal cancer underwent volumetric modulated Arc therapy.Radiother Oncol.2024;199:110438.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Chen L, Ouyang Y, Liu S, Lin J, Chen C, Zheng C, et al.Radiomics analysis of lymph nodes with esophageal squamous cell carcinoma based on deep learning.J Oncol。2022;2022(1):8534262.
Yang M, Hu P, Li M, Ding R, Wang Y, Pan S. Computed tomography-based Radiomics in Predicting T stage and length of esophageal squamous cell carcinoma.前Oncol。2021;11: 722961. 2021.
Du KP, Huang WP, Liu SY, Chen YJ, Li LM, Liu XN, et al.Application of computed tomography-based radiomics in differential diagnosis of adenocarcinoma and squamous cell carcinoma at the esophagogastric junction.世界j胃肠道。2022;28(31):4363.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
致谢
Authors are grateful to the Researchers Supporting Project (ANUI2024M111), Alnoor University, Mosul, Iraq.
道德声明
道德认可
The need for ethical approval was waived off by the ethical committee of Alnoor University, Nineveh, Iraq.This study was conducted in accordance with the Declaration of Helsinki.
Consent to participate
It was waived off by the ethical committee of Alnoor University, Nineveh, Iraq.
竞争利益
作者没有宣称没有竞争利益。
附加信息
Publisher’s note
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。
权利和权限
开放访问This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material.您没有根据本许可证的许可来共享本文或部分内容的改编材料。The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visithttp://creativecommons.org/licenses/by-nc-nd/4.0/。重印和权限
Cite this article
Alsallal, M., Ahmed, H.H., Kareem, R.A.
等。A novel framework for esophageal cancer grading: combining CT imaging, radiomics, reproducibility, and deep learning insights.BMC Gastroenterol25 , 356 (2025).https://doi.org/10.1186/s12876-025-03952-6
已收到:
公认:
出版:
doi:https://doi.org/10.1186/s12876-025-03952-6