增强现实显微镜以弥合AI和病理学家之间的信任
作者:Karasarides, Maria
介绍
数字病理学中深度学习和生成人工智能(AI)应用的加速性是变革性的,并且可能优于典型病理实验室的准确性和可重复性基准1。新兴生物医学AI模型的著名例子包括检测有丝分裂图2,,,,3和肿瘤增殖4,前列腺癌分级5,淋巴结转移检测6和预测预测7,,,,8,,,,9,,,,10,除其他应用11。特别引人注目的是在视觉图像分析和自然语言文本接受培训的机器学习基础模型12,,,,13最近的进步旨在生产将患者病历与组织学结合结合的高性能模型14,,,,15。但是,现实世界中的AI病理系统的采用大大落后,并且越来越多地定位病理学家来处理黑匣子输出,这些输出在常规临床实践中引发了有关AI可靠性的问题16,,,,17。因此,在实践病理学家手中建立AI系统的可信赖性是为了实现患者护理的改善而需要的。
在AI和病理学家之间建立信任的需求在诊断免疫组织化学(IHC)方面特别高,这种方法受到了两个关键挑战的负担:(1)在诊断IHC行业中缺乏分析参考标准,以及(2)对主体染色的重大依赖于对手动评估的方法来确定,该方法是由核对方法来确定的。生物标志物的分析浓度1,,,,18,,,,19。因此,诊断IHC没有明确的绝对真理,必须依靠观察者间的协议作为评分可靠性的估计。虽然IHC伴侣诊断和相关评分方法的监管部门批准是基于对注册试验中临床反应的严格验证,但现实世界的实施IHC测试的前载有变量,并且对生物标志物积极性的变量进行了广泛的评估。18,,,,20,,,,21。重要的是,这种缺乏地面真理对于训练和微调AI模型也有问题,因为注释的训练集源自病理学家对物理IHC污渍的主观解释,而不是由普遍参考标准保护的基于决策规则的标准。
此外,较差的活检抽样,异质组织学,复杂的评分方法和分类阳性截止性为定性带来了难度,充其量是诊断IHC患者选择的半定量性质。对于PD-L1,用于检查点抑制剂免疫疗法的预后和预测性患者选择生物标志物,四个FDA批准了PD-L1 IHC套件22,,,,23,,,,24,,,,25,,,,26,,,,27可商购,多种PD-L1抗体用作实验室开发的测试(LDTS)。最初,蓝图项目28,,,,29提供了跨分析的一致性估计值,但最终在指导现实世界PD-L1测试协调方面无效。从那时起,大量特定的一致性研究继续使核心挑战尚未得到解决,但强调了复杂的评分方法,例如合并的正分数(CPS)和唯一的PD-L1阳性临界值对病理学家来说是有问题的30,,,,31,,,,32,,,,33,,,,34,,,,35。
尽管面临这些挑战,但IHC测试是一种病理学主力,旨在提高测试可重复性和定量准确性的重新努力。免疫组织化学中的分析标准化联盟启动了工作,以定义IHC敏感性阈值,用于开发IHC参考材料和标准化校准器36乳腺癌的第一届乳腺癌IHC参考材料FDA清除了临床用途37,,,,38,,,,39,,,,40。同时,医疗保健利益相关者正在认识到实施AI功能并寻求确定整合杠杆的影响,而病理学家则处于临床决策者的独特地位,敏锐地意识到IHC陷阱,正在努力应对自动化AI模型的可信度及其在工作过程中的信任度41,,,,42。
为了了解塑造病理学中AI可信度的因素,我们设计了一个框架,该框架通过我们的PD-L1 CPS AI模型实现了增强现实显微镜(ARM),并测试了手动和AI-AI-Asissance评分PD-L1 CPS的比较效果。ARM-AI系统使我们能够利用熟悉的载玻片工作流程,同时通过确保在相同条件下运行的病理学家(包括评估相同的视野(FOV)),使用统一的显微镜设置和相同的得分时间来降低评分可变性的来源。
结果
多器官基础检测模型
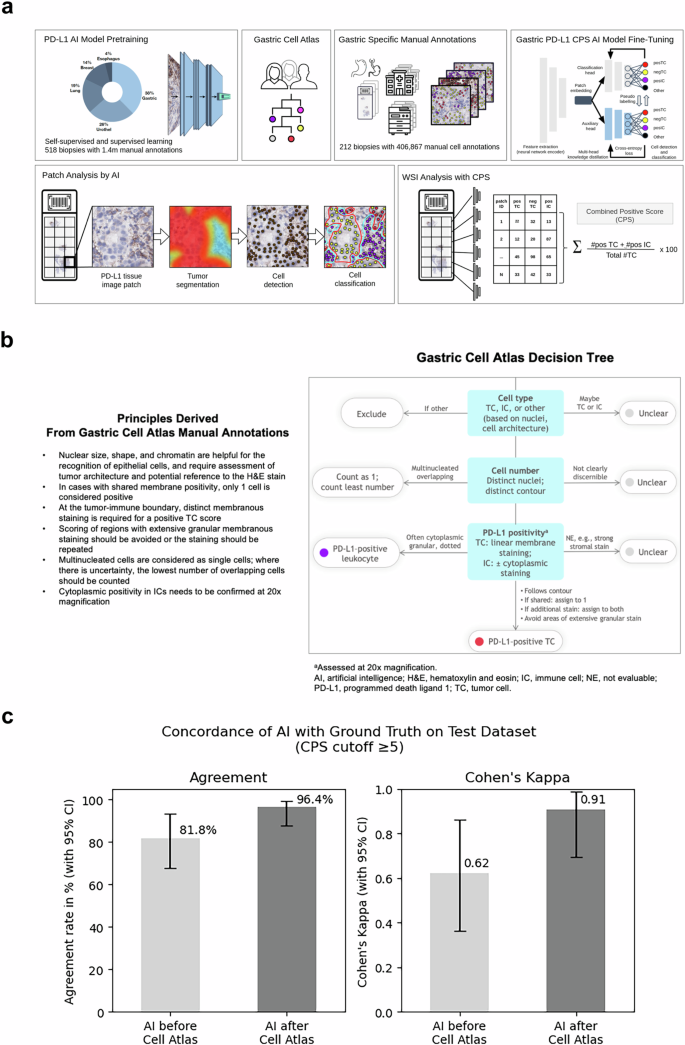
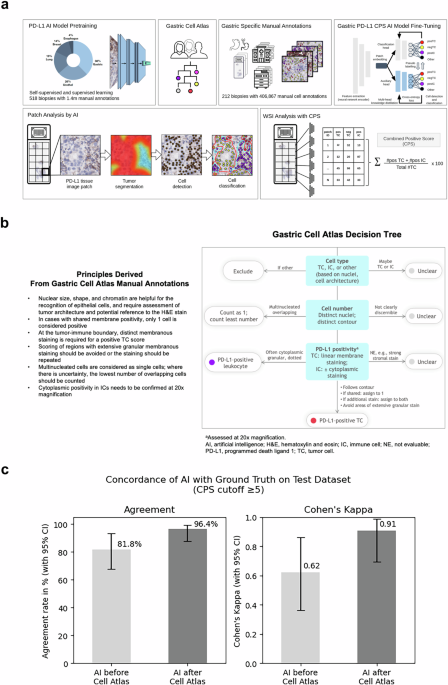
遵循大语言模型的一般研究趋势,AI基础模型目前正在主导计算病理研究空间12,,,,15,,,,43,,,,44,,,,45,,,,46,,,,47。现有的病理AI基础模型使用斑块级特征提取并不一定要明确学习有关细粒细胞和亚细胞结构,即使这是精确检测细胞,细胞膜,亚细胞染色和细微的组织分割的关键。我们使用深层神经网络建立了基于细胞的多器官IHC基础模型,用于需要检测细粒的组织学结构的临床任务。采用混合学习方法,我们结合了自我监督的学习,以检测一般的组织结构和监督学习,以检测具有重要医学相关性的显式生物细胞结构。使用多头知识蒸馏,我们实现了模型正则化和鲁棒性。为了进行监督的学习,我们使用了各种器官的手工制作的细胞注释和细微的组织分割结构。从16个机构中得出的多个器官中的518例多器官活检病例中利用140万个注释(图。1a),我们的AI模型能够检测单个细胞,分类为肿瘤或免疫,并检测PD-L1染色阳性。我们强调使用异质数据的重点是采用战略性的,以补偿染色可变性和扫描仪/显微镜多样性的差异,以实现临床级AI所需的鲁棒性。
胃PD-L1 CPS AI模型。开发了一个多器官基础IHC AI模型(Mindpeak,汉堡,德国),并改编了GC/GEJC/EAC活检组织中CPS方法检测PD-L1的表达。对神经网络(NN)PD-L1 AI模型的预审进,采用了140万个注释,该注释来自来自16个实验室的518例多器官活检病例。通过构建PD-L1 CPS特定决策树(胃细胞地图集)来调和异质组织学和复杂特征,从而完成了细胞注释的细化。胃特异性手动注释由212 GC/GEJC/EAC活检病例(28-8 IHC染色的WSI)确定,该病例来自11个实验室,7个扫描仪,包括406,867 GC/GEJC/EAC细胞。通过使用多头知识蒸馏进行胃PD-L1 CPS AI模型的填充。逐步采用斑块分析来鉴定PD-L1组织图像斑块,然后识别肿瘤分割(红色浸润性肿瘤,与蓝色肿瘤相关的免疫细胞),然后细胞鉴定和最后的细胞分类。对于CPS,定义了PD-L1阳性TC(Postc,Red),PD-L1负TC(NEGTC,黄色)和PD-L1阳性IC(posic,Purple)的单细胞分类。总肿瘤细胞(TC)是根据Postc加上NEGTC计数计算的。CPS等于Postc的数量,加上姿势的数量,除以总可行TC,乘以100,在所有情况下都是数学得出的。WSI通过胃PD-L1 CPS AI模型作为最终输出分析。b胃细胞地图集。在PD-L1染色解释中具有深厚专业知识的三位病理学家回顾了31例GC/GEJC/EAC病例,具有挑战性的组织学和复杂的细胞特征,包括至少以下标准之一:重叠的细胞;微弱,共享和/或颗粒状染色;肿瘤相关的单核炎症细胞的存在;不可见作的肿瘤或非肿瘤细胞;存在或不存在肿瘤侵袭性;并准确地量化细胞数量。设计了针对细胞类型,细胞数和PD-L1阳性的决策标准。总结了胃细胞图集手动注释的原理,并将其与胃细胞的决策树一起提供给参与胃PD-L1 AI模型训练阶段的病理学家。c胃PD-L1 CPS AI模型性能。对55例胃癌病例进行了胃PD-L1 CPS AI模型的初步性能,该病例与训练套件无关,来自3个实验室和3个扫描仪,并用PD-L1 IHC 28-8 PharmDX Assay染色。WSI由2位专家病理学家审查,并分配了CPS¥5 PD-L1参考共识评分。左,胃PD-L1 CPS AI模型在胃细胞决策规则(81.8%)和之后(96.4%)之前对参考共识评分的一致。是的,Cohen的Kappa在胃细胞的决策规则(0.62)和之后(0.91)之前。
我们的AI模型通过首先识别可评估的组织,然后分割肿瘤组织,然后进行细胞检测和分类,然后在所有分析的斑块中汇总(图。1a)。为了在胃癌样品上部署AI模型,我们使用胃,胃食管连接和食管腺癌(GC/GEJC/EAC)活检的AI模型通过CPS方法来检测AI模型来检测PD-L1的表达。Based on cell counts, the CPS methodology requires the identification of PD-L1 on tumor cells and immune cells (lymphocytes and macrophages) with an output that is mathematically derived and defined as the number of positive tumor cells (posTC) plus the number of positive immune cells (posIC), divided by total viable tumor cells (TC), multiplied by 100. Without fine-tuning, our IHC foundation model was empirically在一个测试集上进行评估(n= 55)PD-L1胃癌活检病例用PD-L1 IHC 28-8 PharmDX分析染色,从三个机构和三个不同的扫描仪获得(图。1C)。就案例分类而言,使用CPS的临床相关临界值为正面或阴性,PD-L1参考手册共识评分被视为基础真相。对于每种情况,共识均来自两位病理学家的分数,并在观点差异时通过讨论进行裁决。将IHC基础模型的输出与基于预先指定的截止的共识评分进行了比较,导致协议速率为81.8%(95%CI,68.6%,94.3%)和Cohen S Kappa值为0.62(95%CI,0.36,0.36,0.86)。从单个细胞角度来看,IHC基础模型输出的目视检查揭示了挑战性胃特异性肿瘤组织特征和PD-L1染色阳性的局限性。我们的发现与Robert等人先前的研究保持一致。48当病理学家手动在胃癌活检WSI上手动评分PD-L1 CP时,显示出类似的不一致。
胃癌PD-L1 CPS AI模型的病理学家在环境中进行鉴定
如前所述,众所周知,基于IHC的准确和可重现的生物标志物读数很难获得,这主要是由于分析参数和染色解释可变性。CPS方法尤其有问题,因为它们解释了肿瘤和免疫细胞中的PD-L1染色,并且通常在病理学家中产生高度不一致的输出。这些差异还构成了开发准确的IHC AI模型的挑战,其中需要一致且可靠的培训数据。为了应对这一挑战,我们为困难的胃癌组织架构进行了评分指南,这些指南可以系统地用于病理注释。专家病理学家而不是注释病理学家的直接参与使我们能够汇编有关如何以专家级精度注释组织区域和单个细胞的准确而详尽的决策规则。这些决策规则被赋予注释病理学家,并用于开发微调的PD-L1 CPS AI模型。
我们从31个GC/GEJC/EAC活检案例中确定了不同的ROI,并从AI基础模型训练中产生模棱两可的ROI。每个ROI中的所有单个细胞均由三位专家病理学家独立评估PD-L1 IHC分析。对所有31例活检病例的裁决都进行了亲密的讨论,以得出共识,并就细胞类型,细胞号和PD-L1阳性制定决策规则。由此产生的决策规则总结为胃细胞地图集(图。1B) address areas of difficulty encountered by pathologists and AI models when assessing PD-L1 CPS, including gastric-specific tissue and cell architectures, recognition and distinction of tumor cells and a variety of immune cells, counting of overlapping and multinucleated cells, determination of PD-L1 positivity on tumor cell membranes and number of PD-L1 positive tumor cells in the case of shared membrane positivity.
通过核对异质组织学和复杂的细胞特征,我们采用了胃细胞地图集来指导进一步的注释并实现精制的PD-L1评估,以产生准确的PD-L1 CPS IHC IHC AI模型。212 GC/GEJC/EAAC活检病例来自11个机构,7种扫描仪和406,867个细胞,包括在审查需要PD-L1 CPS读数的胃癌诊断时挑战病理学家的组织区域,用于明所化IHC基础模型,并生产胃PD-L1 CPS CPS AI模型。胃PD-L1 CPS AI模型的初步性能在以前使用的55例胃癌活检病例的相同测试中进行了评估。与原始的IHC基础模型相比,拟南芥PD-L1 CPS AI模型显示,与参考共识的一致性提高了一致性,一致性从81.8%(95%CI,68.6%,94.3%)到96.4%(95%CI,88.6%,100%,100%)(100%,100%)(100%)(95%)p<<0.05,麦克尼玛的测试)和cohen s kappa - 从0.62(95%CI,0.36,0.86)到0.91(95%CI,0.71,1.0)(p<<0.05,引导)(图。1C)。为了验证胃PD-L1 CPS AI模型的性能,我们回顾性地将其与97个GC活检的独立数据集中的12位专家胃肠道病理学家的PD-L1 CPS读数进行了比较48。就一致性相关系数(CCC)而言,来自PD-L1 CPS AI模型的产量与病理学家的相关性更高(0.59; 95%CI,0.49,0.67)高于病理学家手动分数的平均相关性(0.56; 0.56; 95%CI,0.41,0.41,0.41,0.41,0.41,0.41,0.62),并且相比均高于均高度。意义(p<0.001,自举)(补充图。S2)。在PD-L1 CPS€5截止下,PD-L1 AI得分与病理学家手动评分之间的一致性更高(Cohen s- 0.46; 0.46; 95%CI,0.37,0.54)比病理学家之间的一致性一致性(COHEN)(COHEN)(COHEN 9%)0.25%= 0.3.0%。0.48)具有统计显着性(p<<0.05,自举),并且高于原始IHC基础模型(p<0.001,引导)。通过验证两个独立的胃癌活检数据集的AI性能,我们发现与IHC基础模型相比,胃PD-L1 CPS AI模型的性能明显提高,并且与人类PD-L1 CPS IHC读数不属于。信任通过增强现实促进
接下来,我们想研究胃PD-L1 CPS AI模型的影响,作为在载玻片上对PD-L1 CPS评分的病理学家的AI辅助工具。
在常规临床实践中发表的AI算法摄入率低的主要原因之一是在临床级环境中缺乏适当的评估。即使FDA被清除并符合欧洲(CE)认证的AI系统,病理学家也倾向于在批准将整合到实践中之前,可能通过动手经验获得一定的信任。
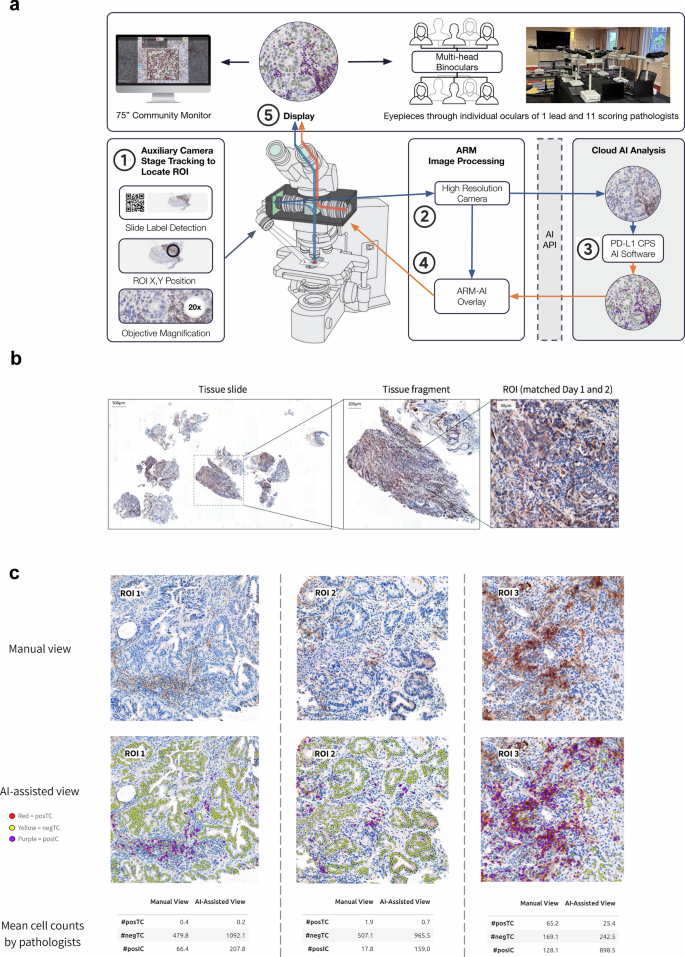
使用ARM-AI系统,我们以受控方式研究了胃PD-L1 CPS AI模型的效用和可信度。十二位专家病理学家,1位未得分样本和11位评分病理学家的主要病理学家参加了为期两天的实验,以实时比较手动与AI-AI辅助评分的35 GC/GEJC/GEJC/EAC活检病例,评估相同的ROI。为此,我们使用Olympus BX53轻型显微镜设计了一个ARM-AI系统,该光学显微镜使用增强的ARM ARM单元进行了改装,该单元连接到本地计算机工作站进行手臂图像处理。通过将16个单位的双眼头系统连接到光学显微镜,可以使相同ROI的多人观看成为可能(图。2)在75英寸监视器上同时显示以进行社区查看。铅病理学家操纵了ARM-AI系统,以确保照明条件是最佳的,并且通过显微镜和数字显示器的图像清晰且清晰。从辅助摄像机启用舞台跟踪的实时进料可以通过X定位每个ROI,Y以20倍的放大坐标确保从第1天到第2天匹配的ROI匹配。PD-L1 CPS AI分析的覆盖层可以由铅病理学家触发,以通过按纽扣按钮显示在FOV中显示的组织中的组织。对ARM-AI系统进行了编程,以将各个组织图像发送到操作胃PD-L1 CPS AI模型的基于云的AI服务器的API。
ARM-AI框架和实验设计的概述。Olympus BX53光学显微镜(显而易见的是,佐治亚州亚特兰大,佐治亚州亚特兰大)的武器单元(Augmentiqs,D.N。Misgav,以色列)与胃PD-L1 CPS AI模型(Mindpeak,Hamburg,Germany)相互作用,并通过16 Binocult vevient veections consection themove of gasters,该单元与胃PD-L1 CPS AI模型(Mindpeak,Hamburg,Dermands)进行了录制。ARM-AI单元连接到计算机工作站,以进行ARM图像处理和基于云的AI分析。社区观看可在75英寸监视器上使用。辅助相机的实时进料使舞台跟踪可以通过X定位ROI,y以20倍的放大坐标。主要病理学家操纵了ARM-AI部门并选择了ROI,但没有参加得分。11个评分病理学家的任务是使用基于Web的数据捕获工具(Mindpeak,Hamburg,Germany)通过单个计算机提交Postc,negTC和Posic的数值;评分病理学家使用手动评分在第1天对35个独特的活检病例进行了评估,并在第2天使用AI助攻。在第2天,正常观看和ARM-AI覆盖层之间的切换由铅病理学家控制,以确保评分病理学家每个样品的等效时间。每个ROI完全包含在第1天和第2 x的单个视野中,y坐标匹配。b数据集的描述。成像以20倍(0.25 mpp)的放大倍率和1 Z平面进行。FFPE原发性肿瘤GC/GEJC/EAC活检的载玻片用H&E和PD-L1 IHC 28-8 PharmDX Assay染色和Robert et al。报告,并报告了基于PD-L1 CPS范围的ROI(零,低,中等,中等,高,高,高)和复杂的Morphology,检查了Robert等人。选择35例活检病例,每个病例中有一个ROI。左,代表性活检显示H&E染色;中间,选择的组织碎片显示案例#34。正确,实际ROI,具有免疫细胞浸润的区域,在第1天手动评估,并在第2天进行AI辅助。c代表ROI是手动评分(第1天)与AI辅助得分(第2天)所见。对于CP,PD-L1 Postc(红色),PD-L1 NEGTC(黄色)和PD-L1Posic(紫色)的单细胞检测。对于每个ROI,计算了11位病理学家的平均细胞计数,显示了AI辅助评分对改善病理学家不熟悉的细胞群体(NEGTC和POSIC)的潜在影响,并且/或或/或改善大量细胞种群的估计(POSTC)。
胃PD-L1 CPS AI模型通过捕获足够大的查看区域(包括外围边缘)以外的核心ROI,分析了组织图像。AI输出由(a)具有坐标的单个单元组成,(b)细胞类型和(c)相应的CPS评分。从眼部看,病理学家能够将PD-L1 CPS AI分析视为叠加在包含组织样品的载玻片上的实时数字覆盖,包括可视化PD-L1阳性肿瘤细胞(PostC,RED),PD-L1负肿瘤细胞(NEGTC,negTC,黄色)和PD-L1阳性免疫细胞(PD-L1负肿瘤细胞)。胃PD-L1 CPS AI模型输出的这种增强现实可视化可以由铅病理学家切换,从而使11个评分病理学家可以将AI辅助视图与原始组织样品的无辅助视图进行比较。病理学家的关键实验任务是独立查看每个预定义的ROI,并在第1天手动产生PD-L1 CPS输出(细胞计数和CPS评分),或者在第2天使用ARM-AI系统启用AI辅助。35个独立的活检病例中的每一个都包含一个预先标记的ROI。拍摄了ROI并记录了坐标,以便评分病理学家可以多次查看每个ROI。要求评分病理学家独立评估所有35个ROI,并提供Postc,negtc和sosic的单个细胞计数。计算每个ROI的最终CPS分数,并提交给病理学家,以达成一致或分歧。如果病理学家不同意派生的CPS评分,则可以肠道检查派生的CP并更改基础细胞计数,而不是直接的CPS。因此,最终的CPS输出(无论是手动还是AI辅助)都由11位评分病理学家中的每一个单独控制。
援助对病理学家表现的影响
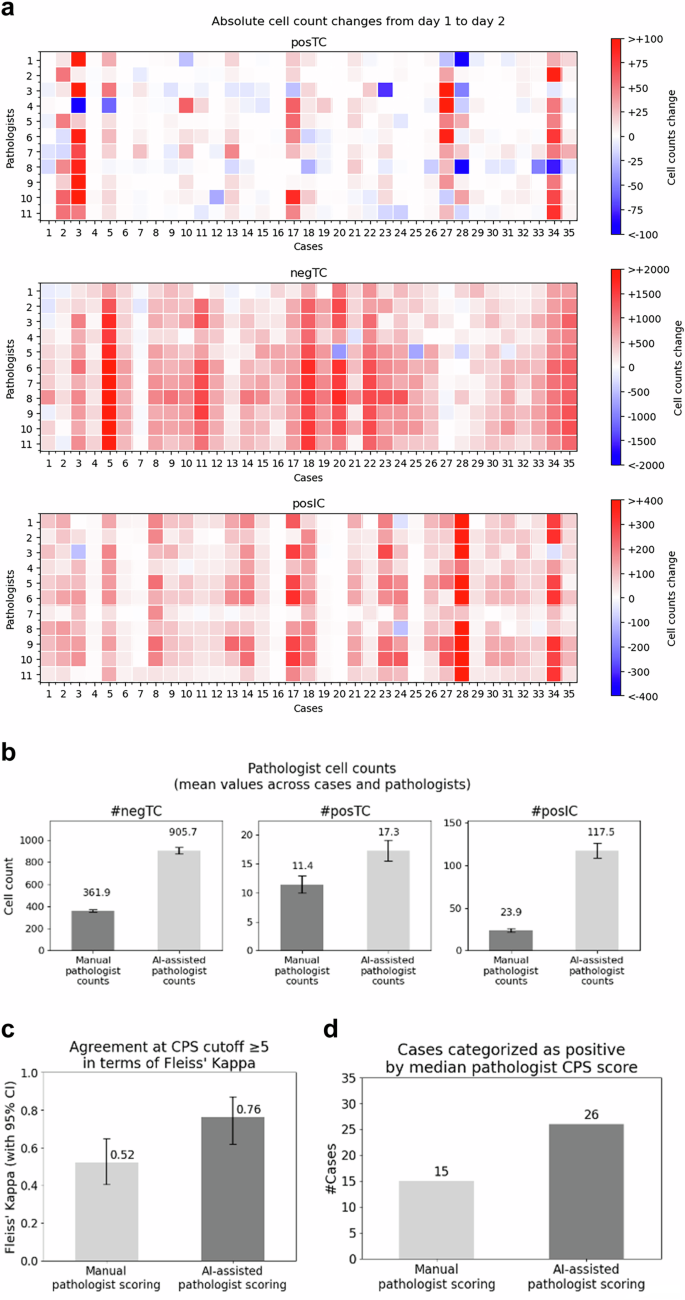
总体而言,AI辅助提高了病理学家对绝对细胞计数的认识,这很可能是通过改善其视觉感知来查看FOV时的意识。Postc,nengTC和posic细胞计数表明,与第1天相比,病理学家报告了在第2天的AI辅助的更多细胞(图。3a),统计学上的显着增加(pnegTC的约2.5倍,postc的1.5倍,姿势为4.9倍(图。3b)。值得注意的是,在第二天,病理学家定性地观察到AI辅助评分促使他们意识到每个ROI中的实际肿瘤细胞的实际数量。
通过细胞类型的11个病理学家中手动与AI辅助评分动态的比较。每个正方形代表11个病理学家中的每一个,分数从手动(第1天)到AI辅助(第2天)的变化,颜色渐变代表了绝对细胞计数,其中蓝色/红色表示第2天较少/更多的细胞。热图:top,top,postc的变化;中间,nengtc的变化;底部,姿势变化。bAI辅助评分提高了对绝对细胞计数的认识,并提高了病理学家的视觉感知的忠诚度。按细胞类型计算的绝对细胞计数:左,negTC手册361.9(349-375)AI辅助905.7(878-933);中间,后C手册11.4(10-13)AI辅助17.3(15-19),右侧手册23.9(22-26)AI辅助117.5(109-127)。cFleissâkappa评估的PD-L1CPS€5的协议。通过使用AI辅助得分Fleissâkappa0.76(0.62-0.87),使用手动得分0.52(0.40-0.65)在第1天对病理学家之间的一致性分析。dAI辅助评分促使病理学家确定11例其他病例为PD-L1 CPS€5。共识CPS€5的计算是从11位个人病理学家的中位数得分。手动评分(第1天)通过AI辅助评分(第2天)确定了15例PD-L1 CPS> 5 vs 26病例PD-L1 CPS€5病例,导致阳性病例增加了31%。
类内相关系数(ICC)用于评估病理学家之间在第2天与ARM-AID辅助评分在第2天进行手动评分的一致性。对CPS组件的得分分析表明,在手动得分但在统计上有显着改善时,病理学家之间的一致性较差(公平的一致性)(p<0.01)与AI援助一致。ICC总结在表中1包括0.38(95%CI,0.31-0.45)手册vs 0.90(95%CI,0.85-0.95)AI辅助TC;0.40(95%CI,0.31-0.47)手册vs 0.91(95%CI,0.85 – 0.95)AI辅助为NEGTC;0.27(95%CI,0.22-0.37)手册vs 0.65(95%CI,0.56-0.71)AI辅助用于POSTC;和0.48(95%CI,0.26 - 0.56)手册vs 0.70(95%CI,0.60 0.75)AI辅助用于Posic。分析了临床相关的PD-L1 CPS的观察者一致性。弗莱斯·卡帕(Fleissâkappa)在病理学家之间达成的一致性显示,总体一致性从0.52(0.40到0.65),第1天的手动评分达到0.76(0.62-0.87),在第2天,AI辅助得分具有统计学意义(p<0.01,差异的自举)(图。3c)。最终,使用PD-L1之类的生物标志物用于选择患者,并可能预测对检查点抑制剂治疗的反应。准确且可重复地对患者进行分类对于优化治疗结果和最大程度地减少治疗不良反应至关重要。为此,我们调查了PD-L1 CPS的调用,每次ROI手动和AI辅助率的5个积极性。通过使用来自每个病理学家的所有11个个体分数中的中位数评分来评估PD-L1 CPS€5阳性ROI的数量。AI辅助评分促使病理学家确定11例其他病例为PD-L1 CPS€5阳性(图。3D)。病理学家确定15例PD-L1 CPS - ¥5阳性,在第1天进行了手动评分,而26例则是PD-L1 CPS€5-5阳性在第2天的AI辅助评分阳性,对应于使用AI-Assissance时阳性病例的31%。在使用AI援助的同时,一些病理学家将评估从PD-L1阳性更改为负(见图。S1在补充材料中)。在手动评分时具有明显不一致的一个情况下,AI援助导致所有病理学家之间完全同意将病例归类为阴性。
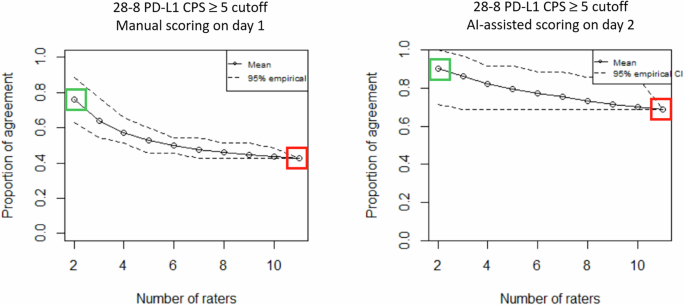
由于没有治疗结果,我们使用评估主观测试所需的观察者(OTEST)评估了发现的重要性30情节分析,该图估计了确定可靠的一致性估计值所需的病理学家数量。图(图。4)表明,需要8-9位病理学家产生可靠的PD-L1 CPS€5估计。比较第1天的手动评分与第2天的AI辅助评分之间的onest阴谋表明,使用手动评分任何两种病理学家对77%的病例一致,而AI辅助评分的任何两个病理学家都同意91%的病例,表明与AI-Assansiscans的同意相当14%。When comparing agreement among all 11 scoring pathologists, agreement was achieved in 43% of the cases with manual scoring in comparison to 69% with AI-assisted scoring, demonstrating a 26% improvement in agreement when using AI-assistance.In summary, higher agreement among pathologists was achieved with the Gastric PD-L1 CPS AI Model, even when viewing was restricted to one 20x FOV.
Overall percent agreement between pathologists on the y-axis vs the number of observers on the x-axis for manual scoring on day 1 (left panel) and AI-assisted scoring on day 2 (right panel).PD-L1 28-8 CPS ≥ 5 cut-off (solid line) and CIs (dotted lines).ONEST analyses indicate that AI-assisted scoring increased overall interobserver agreement.Agreement among any 2 raters using manual scoring is achieved in 77% of the cases (green box left panel) vs 91% of the cases with AI-assisted scoring (green box right panel) resulting in a 14% improvement.Agreement among 11 pathologists using manual scoring is achieved in 43% of the cases (red box left panel) vs 69% of the cases with AI-assisted scoring (red box right panel) resulting in a 26% improvement.
讨论
AI-based diagnostic pathology platforms are delivering unparalleled accuracy in cancer detection and, when fully integrated into real world practice, promise to shorten time to diagnosis and increase accuracy5,,,,49。Developing clinical grade diagnostic IHC AI models for biomarker detection, however, requires a more careful consideration of the parameters used to train the algorithms.IHC biomarker outputs in real-world practice lack quantitative accuracy and assurance that patient level outcomes remain aligned with population level responses observed in clinical trials.Furthermore, there is a gross underappreciation of the complexity pathologists face in routine practice50,,,,51,,,,52。Factors including poor training, high case volume, complex scoring methods, nuanced biomarker cut-offs, biomarker positivity on non-tumor cells, and variable validation approaches have added difficulty to an already arduous task, essentially leaving testing labs and pathologists to shoulder accountability unaided18,,,,20,,,,53,,,,54。Fully aware of IHC discrepancies, pathologists are skeptical about automated AI models and seeking to understand their performance thresholds before fully embracing them as ‘first read’ solutions36。Mindful that future end-to-end AI models should be highly accurate and that human pathologists will retain legal and moral authority over clinical decisions, we sought to understand the effects of establishing trust between AI and pathologists.
In this study, through augmented reality capabilities, we demonstrated that active participation by pathologists in the training and deployment of a novel IHC PD-L1 CPS AI Model, unlike comparatively passive roles as annotators, resulted in mutual performance improvements and facilitated trust with pathologists.We observed that trust could be attributed to (1) allowing human agency, (2) preserving decision accountability, (3) provoking visual perception awareness and scoring behavior and (4) increasing decision confidence associated with difficult cases.Given the potential benefits of fully automated digital pathology systems, we believe that a pathologists-in-the-loop participatory role is a necessary intermediate step that will encourage increased adoption and improved performance of AI models.
Building on a multi-organ cell-based foundation model, our hybrid training approach used both self-supervised learning for recognition of general tissue structures and supervised learning to detect cellular and sub-cellular structures.Our choice of examining PD-L1 CPS ≥ 5 expression on gastroesophageal biopsies was based on our previous work48and other reports55,,,,56,,,,57describing the low concordance rates among pathologists when manually scoring PD-L1 CPS.Given the architectural complexity of gastroesophageal histology and the clinically relevant PD-L1 CPS ≥ 5 cut-off used to determine patient eligibility for checkpoint immunotherapy, we included ROIs with complex heterogeneity that would likely be ignored by both human pathologists and AI models.We first employed direct participation during the fine-tuning process by selecting expert pathologists, qualified as PD-L1 CPS trainers, to examine ROIs that resulted in ambiguous outputs by our PD-L1 CPS AI Model.After adjudication of the ROIs, we constructed a rules-based decision tree termed ‘Gastric Cell Atlas’ and provided it to additional pathologists employed as annotators (Fig.1B)。As a result, our PD-L1 CPS AI Model, trained in total on over 1.8 million annotations from 533 unique biopsies obtained from 16 labs, detected PD-L1 CPS expression on cell structures with 96.4% agreement against a pathologists' derived consensus score (Fig.1C)。To facilitate immersive participation for pathologists, we devised an ARM-AI framework that was operationalized through a multi-head light microscope system accommodating 11 pathologists who were tasked with scoring 35 ROIs on glass slides manually on day 1 and with AI-assistance on day 2 (Fig.2a)。The ARM-AI framework enabled real-time seamless integration of AI into routine workflows using glass slides and leveraged an environment where pathologists could experience AI-assistance to evaluate the trustworthiness of the PD-L1 CPS AI Model outputs.
We found that pathologists’ participation not only improved AI finetuning but significantly improved their own performance, as a group and individually.Agreement, a measure of scoring reliability when ground truth is absent, increased among pathologists across PD-L1 CPS categories with AI-assistance, showing higher ICC for PD-L1 negTC, posTC and posIC (Table1)。The most revealing improvement, however, was in the category of total viable tumor cells.Even within a limited viewing area restricted to one 20x FOV, considered manageable compared to a full WSI, pathologists grossly underestimated the total number of viable tumor cells by at least 60%.Similarly, even though lower in number, they also underestimated the presence of PD-L1 posTC and PD-L1 posIC.Importantly, on day 2 using AI-assistance, pathologists became aware of their tendency to underestimate cell structures, something that they previously ignored or felt had negligible impact.Overall, AI-assistance improved agreement of PD-L1 CPS ≥ 5 (Fleiss’ Kappa 0.52 vs 0.76, Fig.3C), even though we had assumed that agreement would be much higher given that all the scoring was done within one 20x FOV.Of the 35 ROIs scored, 15 cases were deemed PD-L1 CPS ≥ 5 positive by manual scoring and 26 cases deemed positive with AI-assistance.By extrapolation, the impact of manual scoring variability errors indicates that pathologists may be under calling PD-L1 CPS positivity on gastric cancer biopsies.
Agreement analysis using Observers Needed to Evaluate Subjective Tests (ONEST)30demonstrated that AI-assistance resulted in agreement between any two pathologists on 91% of the cases, and among all 11 pathologists on 69% of the cases.Agreement among pathologists as a measure of scoring reliability of subjective IHC staining interpretation is an imperfect proxy with low concordance rates underscoring the need for accurate AI models in routine pathology practice.Although we designed our experiment to achieve as much agreement as possible, by restricting cell counting to one 20x FOV, our data show that staining interpretation variability is multi-layered and not easily eliminated.The observed increased agreement among pathologists using AI-assistance is likely to be even more relevant when evaluating the whole tissue slide.
Regarding trust, we observed that pathologists needed hands-on familiarity through direct participation to become aware of their own baseline scoring behavior and were themselves surprised by the utility of AI-assistance in helping them accurately capture the contents in the FOV.Furthermore, the ARM-AI framework allowed us to pinpoint that human agency is a key component of decision accountability and that both aspects are critical for establishing the trustworthiness of IHC AI models.Although pathologists readily admit that they already work as a type of ‘black box’ themselves, often not able to systematically break down their decision steps, they are reluctant to accept AI tools based on published research without hands-on familiarity42。They want to understand how the AI model produces outputs and evaluate the AI model’s usefulness in their practice42。We noted similar sentiments in our group of 11 scoring pathologists, with acceptance of the IHC AI model when appropriately validated and FDA approved but not on published data alone.Our pathologists unanimously agreed that evaluation of PD-L1 expression on non-tumor cells remains a difficult task, and that pathologists generally score PD-L1 expression with low confidence.
我们的研究有几个局限性。First, the nature of the ARM-AI framework limited our analysis to one ROI per biopsy case.The ROIs were selected from 35 unique biopsy cases that were previously evaluated in a separate study where we documented poor concordance among pathologists when scoring PD-L1 CPS manually48。Since we could not pinpoint the root causes of variability through concordance approaches, even on seemingly straightforward tasks such as viable tumor cell count, we opted to design an experiment that would allow us to evaluate pathologists’ scoring behavior against the PD-L1 CPS AI Model and simultaneously gauge the AI model’s trustworthiness.By limiting our evaluation to one ROI per biopsy, we eliminated the multiple sources of variability present when pathologists score WSIs.When calling PD-L1 positivity, however, pathologists sign out cases based on their holistic evaluation of the entire biopsy section not just specific ROIs, no matter how relevant an individual ROI might appear.Second, our study relied on subjective concordance estimates comparing manual vs AI-assisted scoring outputs.While not optimal, concordance is still the key methodology by which IHC scoring reliability is evaluated since ground truth is not available.While most validation studies evaluate concordance between 1-3 pathologists, our study employed 11 pathologists to ensure we sufficiently captured the variability associated with subjective tests.Ultimately, validation against clinical responses may be needed to overcome the absence of absolute ground truth, however, formal validation of the PD-L1 CPS AI Model nor evaluation of its clinical utility were within the scope of our experiment.Instead, we set out to observe how pathologists interacted with AI and gain insight on the levers of trust and adoption.Finally, the classification of cells as tumor and non-tumor was based on morphological features and did not use specific IHC parameters such as keratin expression.Although this mimics clinical practice, the positivity of misclassified cells as tumor or non-tumor cannot be excluded, particularly challenging for cells with signet ring morphology.
New AI-assistance tools, including our PD-L1 CPS AI Model with integrated ARM, must undergo extensive clinical validation across relevant indications and all available cut-offs (e.g. PD-L1 CPS ≥ 1, ≥5, ≥10) to ensure robust accuracy in routine clinical practice applications.ARM specifically allows the pathologist to more directly interact with annotations, measurements, and AI overlays by significantly enhancing visualization capabilities.Such tools need to be seamlessly integrated within existing pathology infrastructures.Initially, pathologists may resist adoption because they might not be sufficiently familiar with the technology or because they carry a healthy skepticism toward AI models.Hands on experience is crucial for building trust and maturing the technology sufficiently to navigate regulatory standards.Ultimately, the lessons learned can be translated to applying the AI models directly on digital images.
In summary, our findings frame a potential roadmap for building trustworthy IHC AI models and lay out the initial design features of an on-demand digital pathology assistant that can be integrated into real-world pathology workflows.The impact of our work lies in two key areas.The first is to bridge fully automated and fully manual scoring using augmented reality approaches that can be deployed on systems familiar to pathologists.We found that this is a necessary intermediate step to increase trust and adoption as it builds the pathologists' experience with AI and simultaneously improves the performance of pathologists and IHC AI models.The second is to fuel a shift into increasingly quantitative IHC methods that can open the door to truly predictive biomarker discovery as precise clinical decision tools to match patients to therapies.
方法
Gastric Cell Atlas for AI development
A single-cell annotation guideline was established to recognize and annotate both tumor cells (TC) and immune cells (IC) accurately.31 ROIs were analyzed from 31 unique gastric cancer specimens using a 20x magnification.To avoid fatigue and ensure feasibility for pathologists, the number of ROIs was determined by what was assessable in terms of individual cells within a day.Tissues were procured from three different institutions to get diversity in clinical samples: Institute for Hematopathology Hamburg, Hamburg, Germany;((n = 12);Discovery Life Sciences, Kassel, Germany;((n = 10), and Ziekenhuis Netwerk Antwerpen, Antwerp, Belgium;((n = 9).These tissues were stained with the PD-L1 IHC 28-8 pharmDx assay according to the manufacturer’s instructions at three different locations, namely at CellCarta, Antwerp, Belgium;((n = 12);Discovery Life Sciences, Kassel, Germany;((n = 10);and Ziekenhuis Netwerk Antwerpen, Antwerp, Belgium (n = 9).Slides were digitized using either an Aperio GT 450 (Leica Biosystems, Wetzlar, Germany), or a PANORAMIC 250 Flash (3DHISTECH, Budapest, Hungary) scanner.ROIs were chosen by S.B., K.D., and P.F.and contained at least 10–30 cells.The range of 10–30 cells was chosen to prevent fatigue.ROIs were selected as representative regions where single-cell level scoring was difficult due to one or more of the following five criteria: (1) interpretation of faint, shared, and/or granular staining;(2) inclusion of tumor-associated mononuclear inflammatory cells;(3) discrimination of tumor from non-tumor cells;(4) presence or absence of tumor invasiveness;(5) interpretation of cell numbers.
To ensure accurate classification of individual cells, three pathologists with experience in PD-L1 staining interpretation (S.B., J.R., H.S.) convened at Mindpeak, Hamburg, Germany to establish gastric cancer PD-L1 CPS staining interpretation decision rules.T.L., F.F., P.F., G.L.K., M.K., and K.D.were present and participated in the staining interpretation discussion, with final decisions approved by S.B., J.R., and H.S.The study was conducted in two phases.During phase 1, the pathologists were individually presented with the 31 ROIs in consecutive, randomized order via a web-based annotation software (Mindpeak, Hamburg, Germany).The ROIs were independently reviewed in-person by the three pathologists and individual cells were classified into 5 categories (positive/negative TC, positive/negative IC, and unclear).Fibroblasts/endothelial cells were excluded.Pathologists used a recommended stepwise process for identifying single cells for AI scoring in GC, as previously described by Rüschoff et al.58。The three pathologists used identical equipment, under the same conditions.The pathologists had an overview of the whole biopsy slide and the regional tissue context in the tool.To simulate an AI algorithm analyzing the IHC slides, the pathologists were asked to classify cells based on IHC images only.During phase 2, the three pathologists gathered in front of a large computer monitor displaying their individual solutions for each ROI from phase 1 to discuss these.The solutions were presented in a randomized and anonymized manner to avoid bias.The pathologists discussed ambiguities in the existing annotation processes, analyzing all cells that were not unanimously classified in phase 1. Discrepancies were resolved by consensus, and output was used to inform the roadmap.With especially difficult cases, pathologists were allowed to request corresponding H&E-stained images to resolve queries, but this was only used in six instances to assess the extent of immune cell infiltration into the tumor.The pathologists jointly defined annotation principles and rules and decisions were noted by the study team.A consensus solution was developed for each ROI and was recorded separately by the study team under the pathologists’ supervision.The pathologists reviewed the guidelines formulated in the discussion to ensure their general applicability and incorporation in the single-cell annotation roadmap presented here.The resulting Gastric Cell Atlas contained rules for the following aspects: (1) Positivity Interpretation: Scoring specimens with extensive granular (non-linear) membrane staining should be avoided.If most of the membrane staining is granular, then the specimen should be considered as non-assessable.In case of difficulty differentiating light brown from gray tumor membrane staining, clearly negative tumor cells in the surrounding area can be used as control/reference.For tumor and immune cells expressing both cytoplasmic and membrane staining, any strong linear membrane staining that can be distinguished from cytoplasmic background should be considered as positive.Immune cells with faint borderline membrane/cytoplasmic staining should be reassessed at lower magnification (equivalent to 20x).Not only positive and negative tumor cells, but also positive and negative immune cells should be annotated.If positivity or cell class cannot be determined unanimously, a cell should be marked as “unclear†and be excluded from assessment.The principle of assessing relevant cell classes could be generalized to all membrane biomarkers and all required cell classes.(2) Shared Positivity: When two or more tumor cells (or tumor and immune cells) share convincing partial linear membrane positivity, only the cell with staining following the contour of the cell membrane is to be considered positive.Stromal staining in rare foci can be overwhelming and can obscure the cellular outlines and characteristics.Tumor or inflammatory cells staining in these regions is not to be counted as it is not cell specific.(3) Tumor vs Non-Tumor Classification: Both cellular and nuclear morphology and architectural context enable a good distinction of invasive viable tumor cells from non-invasive/non-tumor cells in PD-L1 scoring.Nuclear size and shape are not always reliable criteria particularly when the tumor cells are small and have indistinct nucleoli.Plasmacytoid tumor cells can also be difficult to assess.In such cases, other morphological criteria and surrounding context should be considered.Additionally, pathologists can rely on the H&E stain to distinguish epithelial cells (normal form and tumor) as well as stromal cells (immune and non-immune).Since this may not be possible, however, we chose to mark a cell as “unclear†if its class could not be determined.Such cells can then be excluded in the training data for AI development, resulting in less ambiguous training and increased model stability.Particularly, “unclear†cells were excluded for the development of our Gastric PD-L1 CPS AI Model.(4) Invasiveness: Any epithelial cells that are part of carcinoma in situ (CIS), glandular non-invasive dysplasia as well as the associated mononuclear inflammatory cells (MICs) were excluded from the score.Histology criteria of invasive cancer include tubule / papillary formation, tumor budding, mitotic activity and nuclear pleomorphism.In regions with borderline criteria, context is to be assessed at a lower magnification and if it is cancer associated it must be considered invasive.5) Cell Count: Epithelial cell maps are helpful for distinguishing individual cells.Care should be taken to ensure that multinucleated tumor and inflammatory cells (i.e. cells with a single contour) are counted as one cell.In the case of overlapping cells, those with one cell-contour (e.g., solitary membrane positivity) should be counted as one cell.Groups of cells with multiple contours that distinctively follow their respective borders are to be considered individual cells.If uncertain, the least number of overlapping cells should be counted.6) Tumor associated inflammatory cells: Tumor-associated mononuclear inflammatory cells (MICs) are directly associated with tumor response and should be assessed using 20x magnification.Any pre-existing non-tumor related inflammations and lymphoid follicles must be excluded.In case of peritumoral retraction artefacts, the positive mononuclear inflammatory cells at both sides of the artefact should be considered (reconstructed) as tumor-associated and therefore included in the score.
PD-L1 CPS AI Model and development
AI software, based on deep learning with convolutional neural networks, for PD-L1 CPS quantification in GC/GEJC/EAC samples was developed at Mindpeak and termed Gastric PD-L1 CPS AI Model.It detects cells in IHC stained tissue, distinguishes CPS-relevant tumor and immune cells from other cells, and subsequently calculates cell counts and CPS scores.For a given image, the software analysis proceeds along a pipeline of image processing steps, including neural network models for tissue segmentation and cell detection.First, tissue is detected and distinguished from background using computer vision techniques based on image gradients and color distributions.Then, the tissue is segmented into invasive tumor, tumor-associated immune cell areas, and other non-CPS-relevant tissue areas, using a neural network.In the next step, using relevant tissue segments, cells are detected and classified into either one of the three cell classes relevant for CPS (posTC, negTC, posIC), or as other, irrelevant cell, using a second neural network.Cell detection is accomplished by predicting the probability of each pixel as part of a cell and then clustering neighboring pixel groups with high probability scores into distinct cell entities.Cell classification is achieved by predicting every pixel in a cell class (e.g. tumor and immune) and assigning detected cells after cell detection to classes.The employed neural network models consist of 25 convolutional layer blocks with rectified linear unit activation functions and batch normalization.The neural networks underlying the AI model had been trained in an AI learning phase before the respective experiments.During the experiments, neural networks models were locked and not adapted.The final Gastric PD-L1 CPS AI Model was developed by finetuning a multi-organ foundation model for PD-L1 IHC assessment.The underlying foundation model had been built prior to this study on a large multi-organ dataset, involving PD-L1 cases from gastric cancer, urothelial cancer, esophageal cancer, and non-small cell lung cancer, using 1.05 million image patches of PD-L1 stained tissue and 1.4 m manual annotations.
The foundation model was finetuned to GC/GEJC/EAC using a large gastric cancer dataset.Data for AI development, both for pretraining and finetuning, did not overlap with the study data.To achieve robustness and consistency across changing preanalytical variables, this dataset involved data from 11 institutions, 7 scanners and 3 microscopes.This dataset was annotated by expert pathologists with cell and tissue annotations adhering to the rules of the previously developed PD-L1 Gastric Cell Atlas.A compendium for classifying individual cells in GC/GEJC/EAC biopsies was compiled by three expert pathologists, with guidelines for difficult assessment contexts.The resulting dataset with gastric-specific manual annotations used for AI development contained 406,867 manual cell annotations, resulting in a total multi-organ dataset with 1.8 m manual annotations.The dataset was randomly split into three partitions on a per case basis: training, validation, and test (70, 10, 20), stratified by institutions, scanners and microscopes.The primary metric for model development was the F1 score for cell detection and classification to balance positive predictive value and sensitivity.Model selection was performed by assessing this F1 score on the validation partition.Finetuning of the AI model to GC/GEJC/EAC was based on training neural networks with gradient descent using the Ranger optimization algorithm.To promote model robustness and generalization to unseen preanalytical contexts, a semi-supervised training approach was chosen including knowledge distillation via multiple neural network heads for cell detection and tissue segmentation.
ARM-AI framework
An Olympus BX53 light microscope (Evident, Hunt Optics & Imaging Inc., Atlanta, GA, USA) was retrofitted with an ARM unit (Augmentiqs, D.N. Misgav Israel).The ARM system was programmed to overlay Gastric PD-L1 CPS AI Model (Mindpeak, Hamburg, Germany) within the eyepiece and onto the current view of the sample in real-time (augmented reality).For full technical details on ARM, refer to Chen P.C., et al.59and the Augmentiqs home page:https://www.augmentiqs.com/digital-pathology-software-applications/。Multi-person viewing of the sample was made possible by attaching a 16-unit binocular head system to the main microscope (Fig.2)。The ARM-AI system was connected to a computer workstation for image processing and cloud-based AI analysis.Real-time images were displayed on a 75-inch monitor for community viewing and simultaneously available for multi-head binocular eyepieces for the scoring pathologists.Live feed from the auxiliary camera enabled stage tracking to locate ROI through x/y coordinates at 20x magnification.The lead pathologist maneuvered the ARM-AI system.
Experimental study design
GC/GEJC/EAC biopsy samples on glass slides were previously stained with H&E and with the PD-L1 IHC 28-8 pharmDx assay according to manufacturers’ instructions48。A single selected and pre-marked 20x FOV on each of 35 GC/GEJC/EAC biopsies on glass slides was used, with difficult to interpret ROIs including ambiguous identification of positively staining stromal cells, faint or variable intensity of staining, and difficulty in distinguishing membranous from cytoplasmic tumor staining.Each ROI was wholly contained within a single 20x FOV.The area was marked with a green Sharpie permanent marker.11 pathologists, with a median of 10 years in clinical practice, were invited to perform PD-L1 CPS scoring on day 1 and day 2 of the experiment.4 of 11 scoring pathologists (R.G., L.T., R.A., H.W.) had participated in a previous manual scoring PD-L1 CPS study48。The study proceeded in two phases.On day 1, pathologists manually scored the sample.Imaging was performed at 20x (0.25 microns per pixel) magnification in a 1 z-plane to capture a minimum of 100 viable tumor cells, following the CPS pharmDx interpretation manual (Agilent Technologies Santa Clara, CA, USA).Pathologists provided an exact numeric value for the three elements that comprise the CPS (PD-L1 posTC, negTC and posIC) into a web-based study software from individual computers (Mindpeak, Hamburg, Germany).The FOV from day 1 was captured by the study software.On day 2, the 35 cases were randomly presented in a different order from day 1, using the same FOVs as on day 1 by x/y coordinate matching.Pathologists examined cells using the same FOV as on day 1, but with AI-assistance.Cell counts and corresponding CPS scores were recorded in the study software.One pathologist (N.S.) noted that one biopsy possibly contained signet ring cells, difficult to visually identify and distinguish from macrophages, that were not verified by keratin staining.This circumstantial observation cannot entirely exclude the possibility that a small number of cells were misclassified.Toggling between normal viewing and the ARM-AI overlay was controlled by the lead pathologist to ensure the scoring pathologists had equivalent time per sample.
道德批准并同意参加
The study was conducted in accordance and compliance with the BMS Bioethics Policy (https://www.bms.com/about-us/responsibility/position-on-key-issues/bioethics-policy-statement.html)。The samples procured from the Mayo Clinic received IRB committee approval (#20-007665) in compliance with the institution’s IRB review process (https://www.mayo.edu/research/institutional-review-board/overview) and conducted in accordance with the Declaration of Helsinki (purchased with funds provided by BMS).The samples procured from Discovery Life Sciences were processed by all applicable EU and US regulations as specified on the company’s website (https://www.dls.com/resource-hub/faqs) and purchased by funds provided by BMS.
统计分析
The intraclass correlation coefficient (ICC) was calculated to assess agreement among pathologists for cell counts for PD-L1 CPS relevant cell classes.The value of an ICC can range from 0 to 1, with 0 indicating no agreement among raters and 1 indicating perfect agreement among raters.Inter-pathologist CPS agreement was analyzed at a cutoff score of 5, representing the current clinically utilized cutoffs in gastric cancer, using agreement rates in percent as well as Fleiss’ kappa.Fleiss’ kappa is a measure for assessing the reliability of agreement between a fixed number of raters when assigning ratings to several items.It can be interpreted as expressing the extent to which the observed amount of agreement among raters exceeds what would be expected if all raters made their ratings completely randomly.The concordance of the AI model with ground truth values in the validation before the study was measured by Cohen’s kappa.Cohen’s kappa is a statistic that measures the agreement between two raters beyond chance, accounting for the possibility of agreement occurring by chance.95% confidence intervals for model performance were calculated using nonparametric bootstrapping with 1000 samples.Differences in means or medians for a continuous variable between two groups were assessed by a pairedt-测试。Categorical variables were compared using the McNemar test.95% confidence intervals for concordance values were estimated by bootstrapping (n = 1000).The change in overall percent agreement (OPA) as a function of the number of observers was visualized using Observers Needed to Evaluate Subjective Tests (ONEST).Briefly, for any combination of pathologists, ONEST plots evaluate OPA using the proportion of tissue samples upon which all selected pathologists agreed.Calculation of OPA for all permutations of 12 pathologists resulted in 479,001,600 combinations, from which 100 were then randomly selected.The OPA was plotted against the number of pathologists, resulting in a graph that descends to a plateau.The plateau begins at the number of pathologists believed to be required to provide realistic concordance estimates.Data analyses and summaries were performed using R (version 3.6.1 under Windows 10) and Python 3.11.A list of R and Python packages used for analysis is available upon request.
数据可用性
The human tissue samples used for these retrospective analyses remain anonymized.Tissue blocks, procured from the Mayo Clinic, were processed, sectioned, fixed on glass slides, scanned and converted to WSIs.Restrictions apply to data availability of anonymized patient data, and are thus not publicly available, owing to limitations imposed by data-sharing agreements with the data providers.In line with agreements and institutional policies, all data requests, whether for raw or processed data collected from glass slides or WSIs, should be made to the corresponding author (M.K.) and will be evaluated according to institutional and departmental policies to determine obligations to intellectual property or patient privacy compliance requirements.
代码可用性
The deep learning framework used for AI development was PyTorch, which is available athttps://www.pytorch.org/。The software used for basic image processing (OpenCV) is available athttps://opencv.org/。The R software for data analysis is available athttp://www.r-project.org, and the Python libraries used for data analysis, computation and plotting of the performance metrics (SciPy, Pandas, NumPy and MatPlotLib) are available athttps://www.scipy.org/,,,,https://pandas.pydata.org/,,,,http://www.numpy.org/和https://matplotlib.org/, 分别。The PD-L1 CPS AI Model used in this study is proprietary software protected by IP and owned by Mindpeak GmbH.The software operating the ARM system used in this study is proprietary, protected by IP and owned by Augmentiqs (D.N. Misgav, Israel).
参考
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology.纳特。临床。Oncol。 16, 703–715 (2019).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Malon, C. D. & Cosatto, E. Classification of mitotic figures with convolutional neural networks and seeded blob features.J. Pathol。inf。 4, 9 (2013).
文章一个 Google Scholar一个
Wang,H。等。Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features.J. Med。成像(Bellingham)1, 034003 (2014).
Paeng, K., Hwang, S., Park, S. & Kim, M. A Unified Framework for Tumor Proliferation Score Prediction in Breast Histopathology.In: Cardoso, M. et al.(eds) Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support.DLMIA ML-CDS.Lecture Notes in Computer Science, vol 10553, (Springer, 2017).
Campanella, G. et al.Clinical-grade computational pathology using weakly supervised deep learning on whole slide images.纳特。医学 25, 1301–1309 (2019).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ehteshami Bejnordi, B. et al.Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer.贾马 318, 2199–2210 (2017).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Beck, A. H. et al.Systematic analysis of breast cancer morphology uncovers stromal features associated with survival.科学。翻译。医学 3, 108ra113 (2011).
文章一个 PubMed一个 Google Scholar一个
Yuan, Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer.J. R. Soc.界面 12, 20141153 (2015).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Saltz, J. et al.Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images.Cell Rep. 23, 181–193 e187 (2018).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Corredor, G. et al.Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer.临床癌症。 25, 1526–1534 (2019).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Litjens, G. et al.医学图像分析中有关深度学习的调查。医学图像肛门。 42, 60–88 (2017).
文章一个 PubMed一个 Google Scholar一个
Lu, M. Y. et al.A visual-language foundation model for computational pathology.纳特。医学 30, 863–874 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Kim, C. et al.Transparent medical image AI via an image-text foundation model grounded in medical literature.纳特。医学 30, 1154–1165 (2024).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Vaidya, A. et al.Demographic bias in misdiagnosis by computational pathology models.纳特。医学 30, 1174–1190 (2024).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Chen, R. J. et al.Towards a general-purpose foundation model for computational pathology.纳特。医学 30, 850–862 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Parikh, R. B., Teeple, S. & Navathe, A. S. Addressing Bias in Artificial Intelligence in Health Care.贾马 322, 2377–2378 (2019).
文章一个 PubMed一个 Google Scholar一个
Varoquaux, G. & Cheplygina, V. Machine learning for medical imaging: methodological failures and recommendations for the future.NPJ Digit Med. 5, 48 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Magnani, B. & Taylor, C. R. Immunohistochemistry Should Be Regulated as an Assay.拱。Pathol。实验室 147, 1229–1231 (2023).
文章一个 PubMed一个 Google Scholar一个
Kujan, O. et al.Why oral histopathology suffers inter-observer variability on grading oral epithelial dysplasia: an attempt to understand the sources of variation.口服。Oncol。 43, 224–231 (2007).
文章一个 PubMed一个 Google Scholar一个
Bogen, S. A. A Root Cause Analysis Into the High Error Rate in Clinical Immunohistochemistry.应用。Immunohistochem.摩尔。吗啡。 27, 329–338 (2019).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Dabbs, D. J. et al.In Support of Magnani and Taylor.拱。Pathol。实验室 148, 11 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
22C3, A. PD-L1 IHC 22C3 pharmDx.((https://www.agilent.com/en-us/product/pharmdx/pd-l1-ihc-22c3-pharmdx-overview, 2024).
28-8, A. Dako PD-L1 IHC 28-8 pharmDx.((https://www.agilent.com/en-us/product/pharmdx/pd-l1-ihc-28-8-overview, 2024).
US-FDA, R.D.S.Ventana PD-L1 (SP263) Assay.((https://diagnostics.roche.com/global/en/products/lab/pd-l1-sp263-assay-ventana-rtd001235.html, 2024).
CE-IVD, R.D.S.Ventana PD-L1 (SP263) Assay.((https://diagnostics.roche.com/global/en/products/lab/pd-l1-sp263-ce-ivd-us-export-ventana-rtd001234.html, 2024).
US-FDA, R.D.S.Ventanaa PD-L1 (SP142) Assay.((https://diagnostics.roche.com/global/en/products/lab/pd-l1-sp142-assay-ventana-rtd001231.html, 2024).
CE-IVD, R.D.S.-.Ventana PD-L1 (SP142) Assay.((https://diagnostics.roche.com/global/en/products/lab/pd-l1-sp142-assay-us-export-ventana-rtd001232.html, 2024).
Hirsch, F. R. et al.PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project.J. Thorac。Oncol。 12, 208–222 (2017).
文章一个 PubMed一个 Google Scholar一个
Tsao, M. S. et al.PD-L1 Immunohistochemistry Comparability Study in Real-Life Clinical Samples: Results of Blueprint Phase 2 Project.J. Thorac。Oncol。 13, 1302–1311 (2018).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Rimm, D. L. et al.A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer.贾马·恩科尔(Jama Oncol)。 3, 1051–1058 (2017).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wang, X. et al.Concordance of assessments of four PD-L1 immunohistochemical assays in esophageal squamous cell carcinoma (ESCC).J. Cancer Res.临床Oncol。 150, 43 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Tretiakova, M. et al.Concordance study of PD-L1 expression in primary and metastatic bladder carcinomas: comparison of four commonly used antibodies and RNA expression.mod。Pathol。 31, 623–632 (2018).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Prince, E. A., Sanzari, J. K., Pandya, D., Huron, D. & Edwards, R. Analytical Concordance of PD-L1 Assays Utilizing Antibodies From FDA-Approved Diagnostics in Advanced Cancers: A Systematic Literature Review.JCO Precis Oncol. 5, 953–973 (2021).
文章一个 PubMed一个 Google Scholar一个
Krigsfeld, G. S. et al.Analysis of real-world PD-L1 IHC 28-8 and 22C3 pharmDx assay utilisation, turnaround times and analytical concordance across multiple tumour types.J. Clin.Pathol。 73, 656–664 (2020).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Keppens, C. et al.PD-L1 immunohistochemistry in non-small-cell lung cancer: unraveling differences in staining concordance and interpretation.Virchows Arch. 478, 827–839 (2021).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Bogen, S. A. et al.A Consortium for Analytic Standardization in Immunohistochemistry.拱。Pathol。实验室 147, 584–590 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Vani, K. et al.The Importance of Epitope Density in Selecting a Sensitive Positive IHC Control.J. Histochem Cytochem 65, 463–477 (2017).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sompuram, S. R., Vani, K., Schaedle, A. K., Balasubramanian, A. & Bogen, S. A. Quantitative Assessment of Immunohistochemistry Laboratory Performance by Measuring Analytic Response Curves and Limits of Detection.拱。Pathol。实验室 142, 851–862 (2018).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Sompuram, S. R., Vani, K., Schaedle, A. K., Balasubramanian, A. & Bogen, S. A. Selecting an Optimal Positive IHC Control for Verifying Antigen Retrieval.J. Histochem Cytochem 67, 275–289 (2019).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Materials, B.C.S.I.R.IHControls and IHCalibrators.((https://bostoncellstandards.com/products/, 2024).
McFadden, B. R., Reynolds, M. & Inglis, T. J. J. Developing machine learning systems worthy of trust for infection science: a requirement for future implementation into clinical practice.正面。Digit Health 5, 1260602 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
King, H., Wright, J., Treanor, D., Williams, B. & Randell, R. What Works Where and How for Uptake and Impact of Artificial Intelligence in Pathology: Review of Theories for a Realist Evaluation.J. Med。Internet Res. 25, e38039 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wang, X. et al.Transformer-based unsupervised contrastive learning for histopathological image classification.医学图像肛门。 81, 102559 (2022).
文章一个 PubMed一个 Google Scholar一个
Kang, M., Park, S. H., Yoo, S. & Pereira, D. S. Benchmarking Self-Supervised Learning on Diverse Pathology Datasets.arxiv, (2023).
Filiot A. et al.Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling.medrxiv https://doi.org/10.1101/2023.07.21.23292757(2023)。
Campanella G, K. R., et al.Computational Pathology at Health System Scale -- Self-Supervised Foundation Models from Three Billion Images.arxiv(2023)。
Vorontsov, E. et al.A foundation model for clinical-grade computational pathology and rare cancers detection.纳特。医学 30, 2924–2935 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Robert, M. E. et al.High Interobserver Variability Among Pathologists Using Combined Positive Score to Evaluate PD-L1 Expression in Gastric, Gastroesophageal Junction, and Esophageal Adenocarcinoma.mod。Pathol。 36, 100154 (2023).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Du, X. et al.Effectiveness and Cost-effectiveness of Artificial Intelligence-assisted Pathology for Prostate Cancer Diagnosis in Sweden: A Microsimulation Study.欧元。Urol.Oncol。 8, 80–86 (2025).
B, F. Pathology Under Pressure: Unraveling the Exodus.在The Pathologist(2024)。
Cohen, M. B. et al.Features of burnout amongst pathologists: A reassessment.学院。Pathol。 9, 100052 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Smith, S. M. et al.Burnout and Disengagement in Pathology: A Prepandemic Survey of Pathologists and Laboratory Professionals.拱。Pathol。实验室 147, 808–816 (2023).
文章一个 PubMed一个 Google Scholar一个
Food and Drug Administration, H. Medical Devices;Laboratory Developed Tests 21 CFR Part 809. (ed. Services, D.o.H.a.H.) (2024).
Diagnostics, F.a.D.A.A.C.List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools).(2024)。
Fernandez, A. I. et al.Multi-Institutional Study of Pathologist Reading of the Programmed Cell Death Ligand-1 Combined Positive Score Immunohistochemistry Assay for Gastric or Gastroesophageal Junction Cancer.mod。Pathol。 36, 100128 (2023).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Liu, D. H. W., Grabsch, H. I., Gloor, B., Langer, R. & Dislich, B. Programmed death-ligand 1 (PD-L1) expression in primary gastric adenocarcinoma and matched metastases.J. Cancer Res Clin.Oncol。 149, 13345–13352 (2023).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Peixoto, R. D. et al.PD-L1 testing in advanced gastric cancer-what physicians who treat this disease must know-a literature review.J. Gastrointest.Oncol。 14, 1560–1575 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ruschoff, J. et al.HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing.Virchows Arch. 457, 299–307 (2010).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Chen, P. C. et al.An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis.纳特。医学 25, 1453–1457 (2019).
文章一个 CAS一个 PubMed一个 Google Scholar一个
致谢
We thank members of Mindpeak and Augmentiqs who participated in discussions and especially Gabe Siegel and Dan Regelman for strategic direction and assistance in integrating the ARM hardware.We thank Michelle H. Choi, John Marchlenski, Charles Reichley and the technical experts at Hunt Optics and Imaging for assembling the multi-head microscope.We thank members of the BMS IT department for providing laptop computers.We thank Kald Abdallah for supportive discussions and insights.Funding was provided by BMS.We thank and especially appreciate the patients who provided their tissue for scientific analysis.
道德声明
竞争利益
S.B., R.A., D.C., R.S.G., R.P.G., A.M.K., X.L., A.Q., R.M., N.S., L.T., H.L.W., received hourly payment for participation on day 1 and day 2. S.B., J.R., H.S., received hourly payment for work on Gastric Cell Atlas.J.R. is a consultant for DLS (co-founder of Targos now part of DLS), Astellas, AstraZeneca, BMS, Daiichi Sankyo, GSK, Merck Sharp&Dohme, Merck KGaA and QUip and co-founder of Gnothis Inc. R.A.is a consultant for BMS, Merck SD, AstraZeneca and Jazz pharmaceuticals.S.B.is a scientific advisor to Mindpeak, an ad hoc advisor to AstraZeneca, Daiichi Sankyo, Ventana-Roche, a speaker for AstraZeneca, Daiichi Sankyo, Agilent (Dako), Ventana-Roche, Merck, BMS, a research funding recipient of NCI R01CA121932, Eli Lilly and Agilent (Dako), is a Susan G. Komen Scholar, and the Director of ICGA Foundation, India.R.A.is the recipient of research funding from BMS, RAPT pharmaceuticals, StandUp2Cancer, Breakthrough Cancer and the NIH.G.K., M.K., were employees and shareholders of BMS, when the study was conducted.M.K.is a scientific advisor to DLS, ReviveMed, PictureHealth and a Board Member of FT3.J.P. is an employee and shareholder of BMS.T.L., P.F., M.P., K.D., F.F.are employees and shareholders of Mindpeak.E.P.is an employee and shareholder of Augmentiqs Medical.
附加信息
Publisher’s noteSpringer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
补充信息
权利和权限
开放访问This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material.您没有根据本许可证的许可来共享本文或部分内容的改编材料。The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.要查看此许可证的副本,请访问http://creativecommons.org/licenses/by-nc-nd/4.0/。重印和权限
引用本文
Badve, S., Kumar, G.L., Lang, T.
等。Augmented reality microscopy to bridge trust between AI and pathologists.npj Precis.Onc. 9, 139 (2025).https://doi.org/10.1038/s41698-025-00899-5
已收到:
公认:
出版:
doi:https://doi.org/10.1038/s41698-025-00899-5