多模式AI预测肥厚性心肌病中心律不齐的死亡
作者:Trayanova, Natalia A.
主要的
心脏猝死(SCD)是死亡率的主要原因,也是全球主要的公共卫生问题。在北美和欧洲,SCD的年发病率为每100,000人50 1001,,,,2,,,,3。心室心律不齐是SCD中普遍的基本机制1,,,,4,,,,5。植入式心脏扭曲器除颤器(ICD)可以有效终止心律不齐的发作,并且在预防性部署时,会降低由于心律不齐而高风险的患者死亡率(SCDA)3。SCDA风险分层的当前范围左心室射血分数(LVEF)<30英寸35% - 主要适用于缺血性和扩张性心肌病的患者。即使在这些人群中,它也没有提供全面的风险评估6,许多ICD接收者没有从疗法中获得任何有意义的健康受益7。
SCDA风险分层在肥厚性心肌病(HCM)的患者中更具挑战性,这是最普遍的遗传性心脏病8,因为它具有异质临床表现和进展9。HCM在全球每20000个个人中每2000个人中有1个,并且是SCDA的主要病因8,,,,10。LVEF的风险分层标准<30英寸35%不容易适用于该人群,因为大多数患有致命性心律失常事件的HCM患者具有高肌动力学的左心室功能。以前为HCM开发适当的SCDA风险分层方法的努力尚未达到所需的准确性或实用性11。研究的重点是确定其他风险因素,例如心电图变化12,,,,13和遗传变异14,,,,15,,,,16,或解释图像以从心脏磁共振(CMR)成像中提取预测特征17,,,,18,几乎没有影响11。此外,个性化的机械计算心脏模型(心脏数字双胞胎19)在预测HCM的心律不齐方面已显示出一些成功20;但是,高计算需求限制了其对患者筛查的效用。最后,尽管已经努力为HCM开发机器学习风险预测指标,但它们的局限性很大,包括缺少成像数据21,,,,22,,,,23,,,,24,缺乏代表性的人群21,,,,25并且缺乏外部验证21,,,,22,,,,23。紧急临床需要一种全面的方法,以预测HCM患者准确地心律失常死亡的风险,因此可以得到充分的保护。
在这里,我们提出了一种创新的人工智能(AI)模型,名为MAARS(用于心室心律失常风险分层的多模式AI),旨在通过使用多模式医学数据来预测和解释患者中SCDA风险。我们评估了HCM患者的两名异质人群中的方法。MAARS使用基于变压器的神经网络来分析电子健康记录(EHR),超声心动图和CMR报告的多模式医学数据,以及对比增强的CMR图像。MAAR的核心是三个单峰分支网络,用于编码特定于模态知识的知识,以及用于融合知识的多模式融合模块,用于预后SCDA风险分层。我们证明了HCM的所有临床风险评估工具,其对各种人口亚组的公平性及其在医疗保健系统中的普遍性,内部交叉验证和独立的外部测试中的公平性。MAAR还使用基于注意力和归因的解释来提高决策透明度和产生信任。Maars凭借其出色的高性能和可解释性,展示了AI辅助决策在改善HCM管理和SCDA保护中提供医疗保健方面的潜力。
结果
研究概述
在这项研究中,我们使用了两名来自地理分开社区的HCM患者的同类,以开发和验证我们的模型。使用约翰·霍普金斯医院(JHH)-HCM注册表(Maryland)的553例患者组成的553例患者(即,晚期增强的CMR(即晚期gadolinium增强(LGE)-CMR)扫描,用于开发和内部五倍的MAARS交叉侵略。为了评估MAARS的可推广性和跨健康系统的可靠性,我们进一步测试了该模型的独立外部队列,由286例HCM患者组成的HCM患者,这些患者曾在Sanger Heart&Vascular Institute(SHVI)-HCM登记局(北卡罗来纳州)中获得LGE-CMR图像。为了建立该队列的临床风险预测准确性的基线水平,我们应用了所有广泛使用的临床工具:2020年美国心脏病学院/美国心脏协会(AHA)HCM指南26,2023年欧洲心脏病学会(ESC)心肌病指南27和HCM风险SCD计算器28。在内部和外部验证期间,我们将MAAR的性能与所有可用的临床工具的性能进行了比较,以证明多模式AI提供的出色预测能力。我们还对MAAR进行了消融分析,以说明包含多模式数据和多模式融合方法带来的预测改善。为了分析MAAR的公平性,我们评估了不同性别和年龄亚组的模型性能。最后,我们解释了MAARS做出的预测,并分析了其从每种数据模式中得出的知识。
病人队列
JHH-HCM注册表(内部)包括在2005年至2015年的Johns Hopkins HCM卓越中心被诊断出患有HCM的患者。患者的平均持续时间为2。86年,而主要的SCDA终点被定义为持续的SCDA终点,是持续性的心脏心动过速或心室纤维纤维纤维化,导致PACIBRILLICA或胃口。在本研究中包括的553名患者中,有19名(3%)在随访期间经历了SCDA。
SHVI-HCM注册表(外部)包括确认HCM的患者,根据标准指南衍生的定义进行诊断26在2015年至2023年之间在Atrium Health SHVI HCM卓越中心看到的,SCDA的主要终点被定义为持续的心室心动过速或心室纤维化,从而导致除颤器休克或抗抗心律加速器,或者是抗抗心变速箱。在这项研究中包括的286名患者中,25例(9%)在随访期间经历了SCDA。
为了提供两个队列的概述并表征它们的差异,我们对表中的内部和外部队列之间的临床协变量进行了比较1。选定的协变量包括ACC/AHA或ESC指南概述的人口统计和风险因素,以及两个同类群体之间具有统计学差异的所有其他协变量。在内部队列(JHH-HCM)中收集的67个临床协变量,用于MAARS(扩展数据表1和2),外部(SHVI-HCM)队列中缺少14个,其中18个在内部和外部队列之间存在显着差异。我们使用数据插补来填写两个同类中的丢失信息(方法)。
MAARS:多模式AI模型
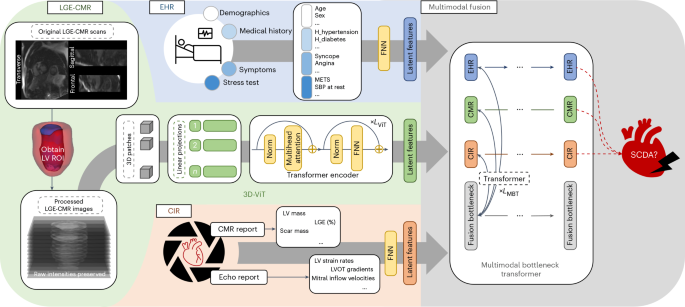
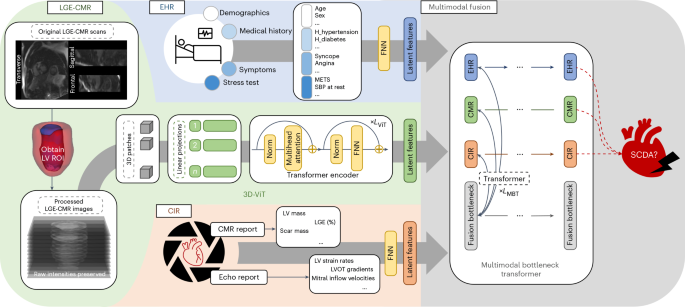
我们的算法(MAARS)通过基于端到端的变压器的设计无缝地整合了不同的输入方式,并协同融合了SCDA风险分层的知识(图。1)。MAARS有三个单峰分支:一个LGE-CMR分支,EHR分支和心脏成像报告(CIR)分支。
MAARS具有三种输入分支的三种输入分支:LGE-CMR图像(左中,绿色),来自EHRS的临床协变量(左上,蓝色)以及CIR的测量值,其中包括CMR和超声心动图报告(左下角,底部,橙色)。处理LGE-CMR图像以获取左心室作为感兴趣的区域,然后用3D-VIT用作输入。EHR和CIR协变量都是结构化的表格数据,并被专用的FNN用作输入。三个输入分支网络的末端连接到多模式融合模块,该模块使用MBT融合知识并学会预测特定于患者的SCDA风险评分(请参阅方法用于详细说明)。回声,超声心动图;ROI,感兴趣的地区;大都会,代谢当量;SBP,收缩压。
LGE-CMR分支网络将具有原始信号强度的LGE-CMR图像作为输入。LGE-CMR图像是多个位置的短轴切片堆叠,可以完全可视化患者左心室的几何形状。图像中的对比度增强区域可视化心肌中疾病诱导的纤维化,该纤维化在机械上创造了心律不齐的底物并导致SCDA。使用原始信号强度的LGE-CMR图像的使用是我们研究的独特特征,因为它保留了扫描中包含的完整,未经处理的信息,并通过避免手动图像解释来消除读取器的可变性。定制的三维视觉变压器29(3D-VIT)用于分析LGE-CMR图像并编码特定于模态知识。在3D数据上学习的固有结构保留了完整的空间信息,并使该模型能够正确地学习LGE-CMR图像和SCDA倾向之间的相关性,从而比限于2D或2.5D透视图的常规模型具有优势。
EHR和CIR分支输入中的协变量反映了该疾病的不同方面。在每个分支中采用了前馈神经网络(FNN)来分析EHR和CIR输入。EHR输入有40个协变量(扩展数据表1)会计患者人口,病史,症状和运动压力测试数据。CIR输入有27个协变量(扩展数据表2)从超声心动图和CMR报告中检索,这些报告使用专家定义的标记来测量心脏功能和形态。值得注意的是,尽管将LGE-CMR图像提供给网络的另一个分支,但包括CMR报告中的四个协变量。通过这种方式,我们确保通过良好的成像功能将专家知识嵌入模型中;这些特征的包含有助于通过模型解释对SCDA预测的贡献进行明确的分析。
在使用单峰分支网络编码特定于模式的知识之后,MAARS使用了多模式瓶颈变压器30(MBT)融合知识并学会预测患者特定的SCDA风险评分。
MAAR优于临床准则
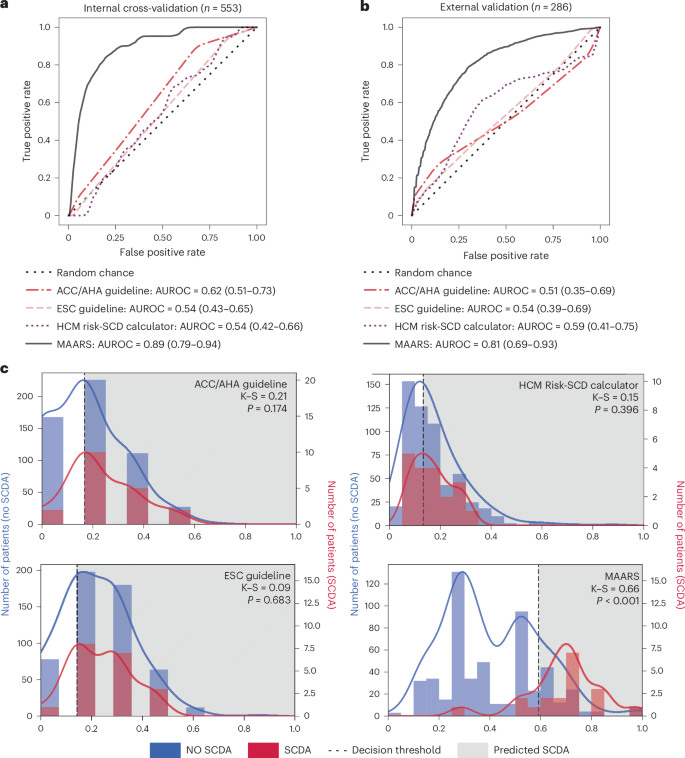
使用接收器操作特征曲线(AUROC),平衡准确性(BA),精确召回曲线(AUPRC),类平衡的Brier评分(BS),灵敏度和特异性(扩展数据表)下的MAARS性能进行了彻底评估。3)。在内部五倍交叉验证中,MAAR的平均AUROC为0.89(95%置信区间(CI)0.79 0.94),平均Ba为0.80(95%CI 0.70 0.89),平均AUPRC平均AUPRC为0.22(95%CI 0.10 0.10 0.34)和0.10 ci 0.16(95%CI 0.16)(95%)(95%)(95%)(95%)(95%)。在外部验证中(扩展数据表4),尽管队列特征的差异和一些缺失的协变量,但MAARS表现出对SCDA事件的强大预测,平均AUROC为0.81(95%CI 0.69 0.93),表明其对不同医疗保健系统的普遍性。对于这两个队列,目前在临床实践中使用的风险评估工具提供了有限的SCDA预测准确性(图。
2)。在内部交叉验证中,ACC/AHA指南列出了七个主要的SCD风险因素,其中有6个分析中包括了六个(左心室根尖动脉瘤缺失),平均AUROC为0.62(95%CI 0.51 0.51 0.73)。ESC指南列出了七个主要的SCD风险因素(此处全部包括),平均AUROC为0.54(95%CI 0.43 0.65)。HCM风险-SCD计算器使用七个风险因素(此处包括此处)来计算5年的SCD风险评分,导致平均AUROC为0.54(95%CI 0.42 0.66)。相比之下,MAAR在AUROC中提供了0.27 0.35的风险评估改善。当将MAARS链接到与临床工具相同的敏感性水平时,它的特异性提高了0.33。同样,在外部验证中,临床工具无法预测SCDA事件,AUROC为0.51 0.59。MAARS在AUROC中提供了0.22的0.30。此外,在内部和外部验证中,MAARS的BS也低于临床风险评估工具(扩展数据表3和4),证明MAAR在预测的概率和真实结果之间具有更好的模型校准。值得注意的是,所有临床风险评估工具和MAAR在外部验证中的性能指标都比内部验证更广泛,这表明绩效估计的不确定性升高,这可能是由于外部患者队列较小。图2:MAARS的性能评估和当前的临床SCDA风险评估工具。一个
由于HCM患者的SCDA事件很少见,因此这项研究中的同伙具有高度不平衡的结果,这对开发有效的风险评估工具构成了巨大的挑战。
我们绘制了从临床风险评估工具和MAAR中的预测风险评分分布,以突出此不平衡数据集中的MAARS性能。我们分析了患者与没有经历SCDA的患者之间的风险评分分布在统计学上存在显着差异(图。2C)。在两个样本的Kolmogorov Smirnov测试中,由三种临床风险评估工具预测的风险评分分布没有显着差异(p> 0.05)在做过和没有经历过SCDA的患者之间。尽管数据集不平衡,但MAAR还是对SCDA风险的歧视性预测,其差异很大(p<0.001)患者与没有SCDA的患者之间的风险评分分布。
多模式融合的优势
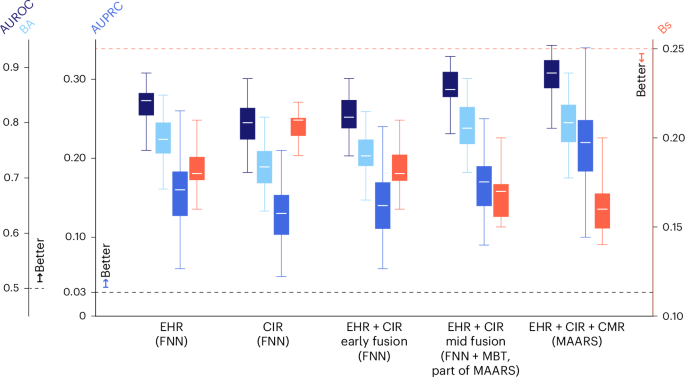
当预测因子中使用单个数据方式时,我们进一步研究了MAARS风险评估绩效的变化。在扩展数据表中提供了三个单峰分支网络(EHR,CIR和LGE-CMR)的性能3。EHR数据反映了有关患者的基线信息,EHR分支网络(FNN)作为基线预测指标,平均AUROC为0.84(95%CI 0.75 0.89),平均Ba为0.77,为0.77(95%CI 0.68 0.85),平均AUPRC和0.16(95%CI 0.16(95%CI)(95%CI 0.06)(95%CI)(95%CI)(95%)。(95%CI 0.16 0.21)在内部交叉验证中。用EHR和CIR进行EHR分支网络(FNN),MAARS以及使用所有三种模式的MAAR进行比较(图。3),我们发现所有性能指标的逐步增强,其CI范围没有差异,这表明在MAARS中融合患者评估的多个方面会减少偏见而不会损害差异。
盒子簇从左至右在x轴是一系列数据模型组合的性能:MAARS内的EHR分支网络使用EHR(EHR,FNN)的临床协变量,MAARS内的CIR分支网络使用CIR(CIR,FNN)的测量(CIR,FNN),FNN,FNN,仅与Ehr和Cir As tufus(Ehr and Cir as forn)(EHR和CIR)的连接(ehr as forn)(ehr)(ehr)(ehr)(ehr's for)(ehr),ehr's for for flousion(ehr)+++++。EHR和CIR(Ehrâ+cir,中期融合,Fnnâ+mbt)和MAARS,具有完整的数据模式(Ehrâ+cirâ+cmr,Maars)。三个蓝色盒子(y左侧的轴)每个集群中的轴代表AUROC(深蓝色,每个集群中左侧的第一个框),BA(浅蓝色,第二个框)和AUPRC(中等蓝色,第三个框),为较高的值表示更好的性能。橙色盒子(y右侧的轴代表BS,对于较低的值表示更好的校准。盒子边界代表第一分位数和第三分位数之间的范围;每个盒子中的白色中心线是平均值;晶须是95%的顺式。所有值都是根据内部队列的交叉验证结果计算的(n= 553)使用自举(方法)。虚线通过随机机会代表基线指标。所有值都是计算出的AUROC,BA,AUPRC和BS。
此外,我们分析了MAAR中多模式融合算法在早期融合模型中的改进(图。3)。早期融合模型是将EHR和CIR数据的串联直接串联的FNN,直接在先前的研究中广泛用于预测心脏不良后果的融合方法17,,,,21,,,,22,,,,23,,,,31。相反,Maars采用了中融合范式(方法)。结果表明,使用EHR和CIR,早期融合模型在交叉验证中的性能较低,甚至比单独使用EHR分支网络的性能低,尽管可以使用更多的数据模式。
跨不同亚组的公平性
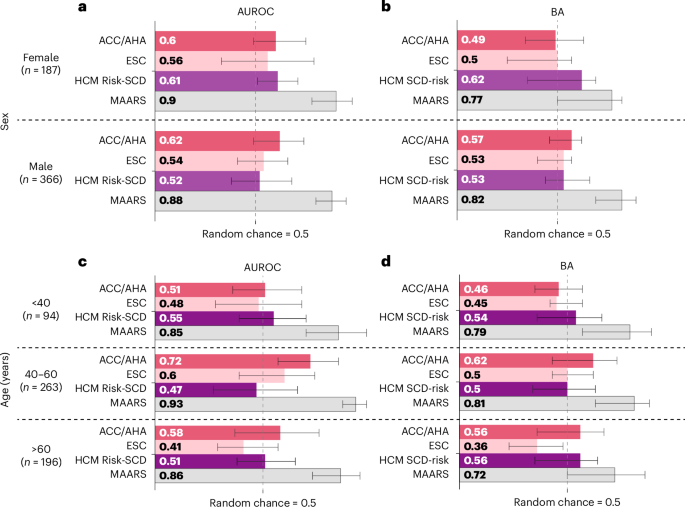
为了评估各种亚组的模型公平性,我们评估了交叉验证中JHH-HCM队列的不同性别和年龄亚组的MAARS表现(图。4)。Maars在每个性别和年龄亚组中的临床工具都远远超过了临床工具。在性亚组评估中,MAARS表现出0.26的AUROC改善0.36,而BA的改善比临床预测指标为0.15 0.29。在年龄亚组评估中,MAAR在中年亚组(40年60岁)的表现最高,AUROC为0.93,Ba为0.81,但在年轻人(<40岁,Aurocâ=aurocâ= 0.85)中的性能略有下降,baâ= 0.85,baâ= 0.79 = 0.79)和(> 60岁),> 60岁,= 0.86 = 7.72 = 7.72 = 76 = <76 = <76 =亚组。在年龄亚组中,MAARS对临床工具的改进甚至更大:AUROC中为0.21 0.46,Ba为0.16 0.36。由于大小有限,对外部队列的公平分析没有进行。
d在不同性别中,显示了多模式AI(MAARS)和临床工具(ACC/AHA指南,ESC指南和HCM风险-SCD计算器)的性能指标(AUROC和BA)(AUROC和BA)(一个,,,,b)和年龄(c,,,,d)亚组。条形长度和条形中的值是手段;晶须是95%的顺式。所有值均基于使用引导(boottrapping)对内部队列的亚组的交叉验证结果进行计算(方法)。
MAAR还表现出预测中的群体间公平性。MAARS性亚组的AUROC变异性为0.02,BA变异性为0.05,AUROC变异性为0.08,年龄亚组的BA变异性为0.09。相反,临床工具具有更高的性能变异性。例如,HCM风险SCD计算器的AUROC高0.09,而女性的BA高0.09。这些是所有模型中最高的变化。ACC/AHA指南在中年亚组中的AUROC可以达到0.72,而Ba为0.62,而它不能预测年龄亚组中的SCDA事件(AUROC = 0.51,BAâ= 0.46),这是SCDA发病率最高的事件。这里提供的证据表明,MAAR可以公平地预测SCDA,并始终超过不同人口亚组中的所有临床工具。
Maars的解释性
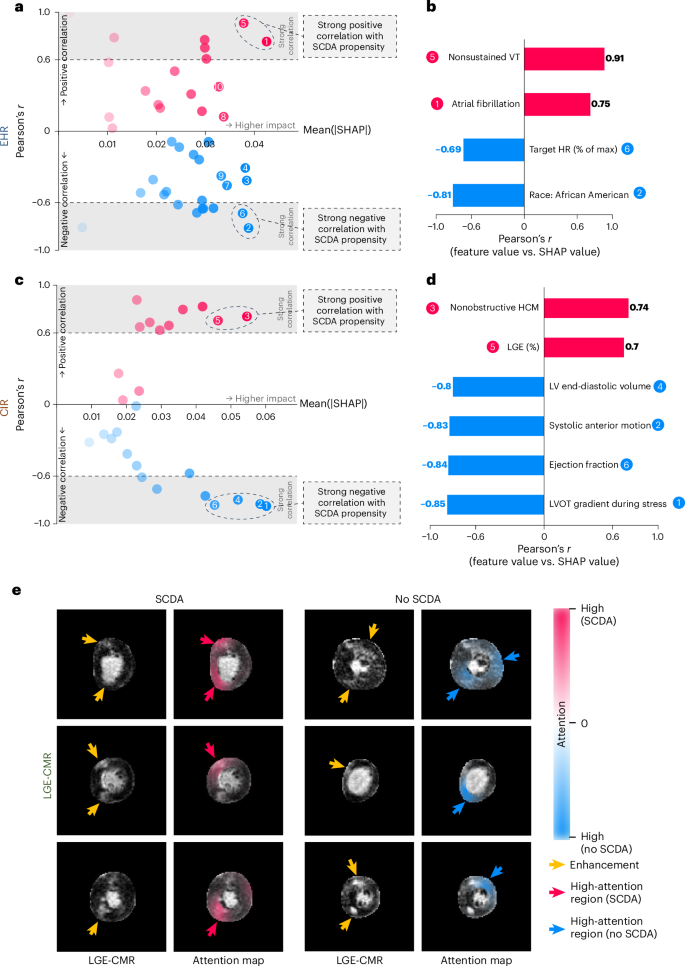
AI算法的解释性对于在临床实践中获得信任和广泛采用至关重要。我们将基于归因和基于注意力的解释应用于MAAR,以解释EHR和CIR协变量和LGE-CMR图像如何有助于SCDA预测。具体而言,沙普利价值32用于量化EHR和CIR分支中每个协变量对SCDA风险评估的归因。协变量的正莎普利值表示协变量有助于患者的SCDA倾向增加,其值量化了影响的大小。使用基于沙普利价值的人口水平的解释,我们确定了最有影响力的(通过绝对沙普利值的平均值衡量)协变量,该协变量也表现出与SCDA倾向的密切相关性(通过Pearson的相关系数衡量(通过Pearson的相关系数衡量)(r)在协变量和沙普利值之间)(图。5)。从EHR分支机构中,我们发现非固定的心室心动过速和心房颤动与SCDA风险增加相关,而在压力测试期间测得的较高的目标心率(%)(反映对运动的年度反应)和非裔美国人种族与SCDA风险降低相关。从CIR分支机构中,发现非目标HCM和更高的LGE负担(%)与SCDA风险增加相关;在压力期间,较高的左心室末端末端体积,二尖瓣的收缩前运动,较高的LVEF和较高的左心室流出道(LVOT)梯度与SCDA风险降低相关。针对个别患者的基于沙普利价值的解释描述了每个协变量对总体风险的增量效应,从而允许决策(扩展数据图2。1和2)。
,从EHR分支输入的临床协变量的内部验证队列中基于Shapley值的解释图。这x轴是绝对沙普利值的平均值(量化协变量的整体影响),并且是y轴是相关系数(Pearson的r)在协变量和沙普利值之间(量化与SCDA倾向的协变量相关)。协变量与r>> 0.6或r<0.6被认为与SCDA倾向有很强的相关性。正相关为红色,而负相关为蓝色。b,模型识别的EHR协变量,影响最高25%(最右边x轴)和与SCDA倾向的牢固相关性。c,,,,d,解释(c)并确定了协变量(d)用于CIR分支。e,基于注意力的LGE-CMR分支网络的解释(留下两列)或没有经历(右两个列)SCDA的患者。第一和第三列中的短轴LGE-CMR图像是LGE-CMR分支的输入的示例,其网络中相应的注意力图正在覆盖LGE-CMR图像,并在第二和第四列中显示。高注意值的区域(对于经历SCDA的患者的红色,未经历SCDA的患者的蓝色)表明,局部像素强度对网络决策过程有很高的贡献。黄色的箭头指向LGE-CMR图像中的对比度增强,红色和蓝色箭头指向高意见区域。人力资源,心率;VT,心室心动过速。
此外,协变量解释分析具有增强模型简化的潜力,这对于临床实用性至关重要。为了进一步分析,我们训练和评估了一个简化的协变量网络(FNN),从EHR和CIR作为输入中最有影响力的协变量(图。5),以及将此协变量网络与现有LGE-CMR分支网络(3D-VIT)和多模式融合模块(MBT)相结合的简化MAAR。这种简化的MAARS模型在内部和外部验证中保持了高度的精度(扩展数据表5),与综合MAARS模型相比,性能仅降低。这突出了模型对不同临床环境的适应性,具有不同的数据可用性。
用于LGE-CMR图像分析的3D-VIT具有先天的注意机制,使我们能够通过突出与网络预测相关的领域来解释图像中收集的知识。我们使用注意力推出量化了受过训练的3D-VIT的注意力流动(方法)并将注意力对输入LGE-CMR图像的关注视为热图,显示图像区域对于预测SCDA风险很重要(图。5e)。在经历过SCDA的患者的LGE-CMR图像中,MAARS通常关注但不限于增强区域的区域。对于没有经历SCDA的患者,LGE周围的MAAR区域对于做出预测似乎很重要。这表明Maars学会了LGE分布与SCDA倾向之间的细微关系。
讨论
在这项研究中,我们开发了一种称为MAAR的AI模型,以通过多种方式使用医疗数据来预测HCM患者的SCDA风险。MAARS具有基于变压器的神经网络结构,包括几个模块:(1)对具有原始信号强度的LGE-CMR图像进行3D-VIT学习,可视化疾病诱导的心肌纤维化分布;(2)用于分析EHR临床协变量的FNN;(3)用于分析标准超声心动图报告和CMR放射学报告的FNN;(4)多模式融合模块MBT,以整体的角度整合知识并分层SCDA风险。我们通过在HCM患者中进行的一系列分析来证明MAAR的表现高,其中我们的模型表明(1)在所有临床工具上,SCDA风险分层性能的显着改善,(2)(2)外部患者群体与来自其他医疗系统的外部患者群体的概括性,在地理位置分开的位置,(3)对不同的患者进行了分组,(3)对不同的clabion cance the Aly Grance the sexii sealsi,(4)(4)(4)(4)(4)。(4)(4)。MAARS凭借其优于现有临床工具的卓越性能,可推广性,公平性和解释性,有可能实质上改善SCDA风险分层和患者护理的交付。
HCM中SCDA的准确风险分层仍然是一个主要的临床挑战,MAARS提供了有希望的解决方案。现有的临床风险评估工具在识别高危患者方面无法获得令人满意的准确性。ACC/AHA的先前临床验证研究26和Esc27指南报告了不同队列的C-指数值不同(从0.52到0.92)33,,,,34,,,,35,,,,36,,,,37。在我们的研究中,我们发现临床风险评估工具具有AUROC(相当于二进制结果设置中的C索引,SCDA与NO SCDA相当于c-Index,这两个同类群体为0.51 0.62,在SCDA风险分层中提供有限的价值。MAAR凭借许多方法上的创新,在内部和外部人群中的所有临床风险评估工具都显着优于所有临床风险评估工具。除了强大的预测性能外,医疗保健系统的普遍性以及人口亚组之间的公平性也突出了MAAR的优势。Supported by MAARS, clinicians can deliver personalized care to their patients and protect them appropriately from SCDA while also reducing unnecessary ICD deployments and the associated drawbacks, namely, infections, device malfunction and inappropriate shocks6。MAARS embodies an important technological innovation in directly analyzing LGE-CMR images with raw signal intensities for SCDA risk prediction.LGE-CMR images carry rich information regarding cardiac pathology but have long been underused due to complexities in image interpretation.
Previous attempts to incorporate LGE-CMR image-derived features24,,,,26,,,,27,,,,38in SCDA risk predictors had a number of limitations, such as manually engineered features being unable to represent intricate anatomical shapes and scar distributions, as well as image interpretations prone to inter-reader variability.Although there have been efforts to automate the CMR image interpretation process with deep learning17,,,,18, raw medical imaging information remains largely underused.Our previous study pioneered learning on LGE-CMR imaging data with raw signal intensities in ischemic cardiomyopathy39using a convolutional neural network;a similar network was used in a recent study25。Here, MAARS excelled in analyzing LGE-CMR images with raw signal intensities by adopting an advanced 3D-ViT network, and we validated the advantage of this approach in two large and diverse HCM cohorts.
MAARS demonstrates the value of integrating multimodality information in clinical decision support systems.Although medical assessments of different aspects of cardiovascular health are routinely acquired, the integration of different data modalities for clinical decision-making has remained a challenge40。Previous studies predicting adverse cardiac outcomes simply built models based on a concatenation of all features17,,,,21,,,,22,,,,23,,,,31;this approach, as used by the reference model shown here, underperforms the multimodal fusion structure in MAARS.The dedicated fusion module using an MBT network in MAARS specifically learns the cross-modality interplay and effectively integrates knowledge.The low performance of the reference model might be due to overfitting caused by increased optimization difficulty from the additional data modality41;however, with the multimodal fusion algorithm, MAARS attenuates the overfitting problem and efficiently uses all data modalities.
The multilevel interpretability is a crucial component of MAARS that distinguishes it from the prevailing black-box healthcare AI methods.The individual-level interpretation not only improves the transparency of AI prediction and, therefore, makes MAARS more trustworthy but also paves the way for precision patient care.From the interpretation of risk prediction for a particular patient, a healthcare provider can identify the personalized factors that lead to high SCDA risk in this patient and potentially prioritize modifying these factors to manage the disease trajectory most effectively.In addition, the population-level interpretation can facilitate the discovery of new knowledge.From the AI interpretation, we observed that some findings align with the current consensus and reaffirm their clinical significance, including nonsustained ventricular tachycardia, higher fibrosis burden (LGE %) and lower LVEF being associated with increased SCDA risk26,,,,27。Conversely, some findings may warrant further investigation.For instance, in this study, systolic anterior motion and higher LVOT gradient were associated with lower SCDA risk;meanwhile, nonobstructive HCM was associated with increased SCDA risk.These findings provide information in the debate regarding the prognostic implication of the presence of LVOT obstruction.Although a considerable amount of evidence42indicates that patients with obstructive HCM are at greater risk of disease-related adverse events, including syncope, heart failure and mortality, recent studies43,,,,44have provided more granularity on the specific clinical outcomes, suggesting that nonobstructive HCM is associated with a higher burden of ventricular arrhythmia events.This contrasts with atrial arrhythmias, which were more common among patients with LVOT obstruction.The pathophysiologic mechanisms behind the higher SCDA risk in nonobstructive HCM might be the greater severity of myopathy45associated with a higher burden of ventricular fibrosis46and the presence of coronary microvascular dysfunction and ischemia47in these patients.However, it should be noted that these AI-derived interpretations reveal statistical associations and correlations rather than establishing causality and, therefore, cannot be directly interpreted as disease mechanisms.This consideration represents a common challenge faced by the broader AI research community.
This study has several limitations and practical considerations that affect the immediate clinical utility of MAARS.First, MAARS was developed on a single tertiary-care center cohort and, therefore, was potentially exposed to institutional or referral bias.While such biases are common in HCM clinical studies, our research benefits from a relatively large internal patient cohort with diverse demographics, disease severity and comorbidities;it is also strengthened by external validation on a cohort from a geographically separate healthcare system.These advantages are particularly valuable given the requirements for comprehensive clinical data and advanced LGE-CMR imaging.Second, despite being robust in a proof-of-concept study, our cohort sizes remain smaller than those of established risk prediction models being used clinically, such as the HCM Risk-SCD calculator28。The limited cohort sizes combined with the rare incidence of SCDA events resulted in a small absolute number of observed endpoint events (19 in the internal cohort and 25 in the external cohort), especially when compared to the complexity of the MAARS model (67 clinical covariates and LGE-CMR images).This created challenges in model development, increased the risk of model overfitting and added difficulty in precisely estimating performance metrics.This limitation was reflected, for instance, in the wide CI in the external validation results, which is wider than the recommendation of CI width ≤ 0.1 (ref.48)。Nevertheless, the event rate observed in this cohort aligns with that reported in other HCM studies28,,,,49,,,,50, supporting the validity of our study design, and MAARS demonstrated statistically significant superior performance over current clinical tools, highlighting the potential value of integrating multimodal data.A third limitation is the lack of detailed time-to-event information, precluding the assessment of the model’s predictive ability over time.Finally, the increased data requirements of MAARS compared to simpler clinical tools may present a practical barrier to its widespread clinical adoption.Collectively, these limitations underscore the necessity for future research to validate the model’s generalizability in larger cohorts and assess its robustness during longer follow-up periods.
Overall, MAARS is a powerful and trustworthy clinical decision support tool for SCDA risk stratification in HCM developed with state-of-the-art multimodal AI technology.It has validated strong performance, fairness and generalizability and provides interpretations for the predictions.MAARS has the potential to substantially improve clinical decision-making and healthcare delivery for patients with HCM, either directly through future integration with automated data extraction systems or indirectly by serving as a valuable proof of concept for the power of multimodal AI in enhancing personalized patient care.
方法
This study complies with all relevant ethical regulations and has been approved by the institutional review boards of Johns Hopkins Medicine and Atrium Health.
Patient population and datasets
JHH-HCM registry (internal)
A retrospective analysis was performed on patient data from the JHH-HCM registry spanning 2005–2015.Enrollment in the registry was based on the first visit to the Johns Hopkins HCM Center of Excellence, where patients meeting the diagnostic criteria for HCM were included.These criteria focused on the presence of unexplained left ventricular hypertrophy (maximal wall thickness ≥15 mm) without evidence of uncontrolled hypertension, valvular heart disease and HCM phenocopies, such as amyloidosis and storage disorders.Patients were followed for a mean duration of 2.86 years (median 1.92 years; 25th–75th percentile = 0.94–4.28 years).The current study focused on a subset of patients with HCM who were enrolled between 2005 and 2015 and had adequate LGE-CMR images, totaling 553 patients (Extended Data Fig.3)。SHVI-HCM registry (external)A retrospective analysis was performed on patient data from the Atrium Health SHVI-HCM registry spanning 2015–2023.
This registry includes patients who presented to the SHVI HCM Center of Excellence with a preexisting HCM diagnosis or were subsequently diagnosed based on cardiac imaging, personal and family history, and/or genetic testing in accordance with current guideline definitions.
Patients within this longitudinal database are still being followed, as the endpoint for registry inclusion is the transfer of care to an outside facility or death.For the purposes of this study, the SHVI-HCM registry was interrogated for patients who had undergone CMR imaging and ICD placement, and enrollment was delineated by the patient’s first visit with the SHVI.
Data collection and primary endpoint
Clinical data, including demographics, symptoms, comorbidities, medical history and stress test results, were ascertained during the initial clinic visit and at each follow-up visit.Rest and stress echocardiography and CMR imaging were performed as routine components of clinical evaluation for all patients referred to the HCM centers.For the internal JHH-HCM registry, echocardiography and CMR imaging were conducted before the first clinic visit, with typically 3 months between the imaging assessment and the first clinic visit.For the SHVI-HCM registry, patients typically underwent echocardiography and CMR imaging after the first clinic visit.The full list of covariates used in MAARS can be found in Extended Data Tables1和2。The data were extracted through a manual search of patients’ EHRs.EchoPAC software (GE Healthcare) was used to quantitatively analyze the echocardiogram and compute related covariates.Of note, the internal and external cohorts have distinct patient populations with different demographic characteristics and different levels of risk factors (Table1)。
The CMR images in the JHH-HCM registry were acquired using 1.5-T magnetic resonance imaging (MRI) devices (Aera, Siemens; Avanto, Siemens; Signa, GE; Intera, Phillips).In the SHVI-HCM registry, most CMR images were acquired using 1.5-T MRI devices (Aera, Siemens; Sola, Siemens), and a small proportion of CMR images were acquired using 3-T MRI devices (Vida, Siemens).LGE images were obtained 10–15 min after intravenous administration of 0.2 mmol kg−1gadopentetate dimeglumine.An inversion scout sequence was used to select the optimal inversion time for nulling normal myocardial signal.All images used were 2D parallel short-axis left ventricular stacks.Typical spatial resolutions were in the range of 1.4–2.9 × 1.4–2.9 × 7–8 mm, with 1.6- to 2-mm gaps.
The primary endpoint for the JHH-HCM registry was SCDA defined as sustained ventricular tachycardia (ventricular rate ≥130 beats per min lasting for ≥30 s) or ventricular fibrillation resulting in defibrillator shocks or antitachycardia pacing.Arrhythmic events were ascertained by reviewing electrocardiogram, Holter monitor and ICD interrogation data.The primary endpoint for the SHVI-HCM registry was SCDA defined as device shock, appropriate interventions or out-of-hospital cardiac arrest.
More details regarding patient inclusion, assessment, follow-up, echocardiography and CMR acquisition can be found in previous work23,,,,51。
数据准备
The multimodal inputs to MAARS included LGE-CMR scans and clinical covariates from EHRs and CIRs (Extended Data Tables1和2)。The labels were the outcomes (SCDA or non-SCDA).The preprocessing steps for LGE-CMR scans (described below) aimed to exclude nonrelevant background information and to standardize the CMR image volume for consistent analysis across all patients.We first obtained the left ventricular region of interest using our previously developed and validated deep learning algorithm52。Once each patient’s LGE-CMR 2D slices were processed using this algorithm, all pixels outside the left ventricle were zeroed out, and the pixels within the left ventricle were normalized by the median blood pool pixel intensity in each slice.Finally, the processed slices were stacked and interpolated to a regular 96 × 96 × 20 grid with voxel dimensions of 4.0 × 4.0 × 6.3 mm.
The EHR and CIR data were structured as tabular data.The input features included in the analysis were ensured to have <40% missing values originally;missing values were imputed using multivariate imputation by chained equations (MICE)53。MICE is a fully conditional specification approach that models each input feature with missing values as a function of all other features iteratively.To address the feature mismatch issue between the internal and external cohorts, we used a MICE imputer based on the internal dataset to impute the missing values in both datasets.After the imputation, the EHR and CIR data were standardized using thez-score method, which involves subtracting the mean and dividing by the s.d.of each feature.
Transformer-based multimodal neural network
Modality-specific branch networks
Three unimodal branch networks are included in MAARS, each learning from a specific input modality: a 3D-ViT29for LGE-CMR images, an FNN for EHR data and an FNN for CIR data.
In the LGE-CMR branch, the image vector embeddings我¶are obtained by dividing the original 3D imagex进入nflattened nonoverlapping 3D image patchesx我and following the operations$$\begin{array}{c}{\zeta }_{{\rm{CMR}}}^{\,0}=\left[{z}_{{\rm{cls}}},E{x}_{1},E{x}_{2},\ldots
,E{x}_{n}\right]+{p}\end{array}$$
(1)
在哪里eis a linear projection,zCLSis a classification token (CLS-token) and ‘p’ is a learnable positional embedding to retain positional information.
The image vector embeddings\({\zeta }_{{\rm{CMR}}}^{\,0}\)are then processed by a sequence oflViTtransformer encoder blocks.Each transformer encoder block,\({\zeta }_{{\rm{CMR}}}^{\,l+1}={\rm{Transformer}}\left({\zeta }_{{\rm{CMR}}}^{\,l};{\theta }_{{\rm{ViT}}}^{l}\right)\), consists of two submodules: (1) a multihead self-attention (MSA) module and (2) a two-layer fully connected FNN.
$$\begin{array}{c}{\nu }^{l}={\rm{MSA}}\left({\rm{LN}}\left({\zeta }_{{\rm{CMR}}}^{\,l}\right)\right)+{\zeta }^{\,l}\end{array}$$
(2)
$$\begin{array}{c}{\zeta }_{{\rm{CMR}}}^{\,l+1}={\rm{FNN}}\left({\rm{LN}}\left({\nu }^{l}\right)\right)+{\nu }^{l}\end{array}$$
(3)
where LN is the layer normalization operation.In the final transformer encoder block, the encoded CMR knowledge,ξCMR, is defined as
$$\begin{array}{c}{\zeta }_{{\rm{CMR}}}^{{\,L}_{{\rm{ViT}}}}=\left[{z}_{{\rm{cls}}}^{{\,L}_{{\rm{ViT}}}},{z}_{1}^{{\,L}_{{\rm{ViT}}}},{z}_{2}^{{\,L}_{{\rm{ViT}}}},\ldots ,{z}_{n}^{{\,L}_{{\rm{ViT}}}}\right]={\rm{Transformer}}\left({\zeta }_{{\rm{CMR}}}^{{\,L}_{{\rm{ViT}}}-1};{\theta }_{{\rm{ViT}}}^{{L}_{{\rm{ViT}}}-1}\right)\end{array}$$
(4)
$$\begin{array}{c}{{\rm{\xi }}}_{{\rm{CMR}}}={\rm{LN}}\left({z}_{{\rm{cls}}}^{{\,L}_{{\rm{ViT}}}}\cdot W\right)\end{array}$$
(5)
在哪里wis a learnable matrix.
In the EHR and CIR branches, processed EHR and CIR data are converted to vectors我¶EHR,,,,我¶CIRfed into two FNNs, with outputsξEHR和ξCIRrepresenting the encoded EHR and CIR knowledge.
$$\begin{array}{c}{\xi }_{{\rm{EHR}}}={\rm{FNN}}\left({\zeta }_{{\rm{EHR}}};{\theta }_{{\rm{EHR}}}\right)\end{array}$$
(6)
$$\begin{array}{c}{\xi }_{{\rm{CIR}}}={\rm{FNN}}\left({\zeta }_{{\rm{CIR}}};{\theta }_{{\rm{CIR}}}\right)\end{array}$$
(7)
Multimodal fusion
Following knowledge encoding from the LGE-CMR, EHR and CIR subnetworks, we used an MBT consisting of multiple blocks to fuse the knowledge across modalities.MBT has demonstrated state-of-the-art performance in multimodal fusion tasks and has a light computational cost30。In each MBT block, the unimodal knowledge vectors concatenated with a shared fusion vector,ξfsn, are fed into modality-specific transformers:
$$\begin{array}{c}\left[{{\xi }_{* }^{l+1},\hat{\xi }}_{{\rm{fsn}},* }^{l+1}\right]={\rm{Transformer}}\left(\left[{\xi }_{* }^{l},{\xi }_{{\rm{fsn}}}^{l}\right];{\theta }_{{\rm{MBT}},* }^{l}\right)\end{array}$$
(8)
The fusion vector in layerl + 1 is updated as follows:$$\begin{array}{c}{\xi }_{{\rm{fsn}}}^{\,l+1}={\rm{Avg}}\left({\hat{\xi}}_{{\rm{fsn}},*
}^{\,l+1}\right)\end{array}$$
(9)
The last MBT block outputs a predicted SCDA risk scorepusing the following equation:$$\begin{array}{c}p={\rm{sigmoid}}\left(\left[{\xi}_{{\rm{CMR}}}^
{{\,L}_{{\rm{MBT}}}},{\xi}_{{\rm{EHR}}}^{{\,L}_{{\rm{MBT}}}},{\xi }_{{\rm{CIR}}}^{{\,L}_{{\rm{MBT}}}}\right]\cdot W+b\right)\end{array}$$
(10)
Model training and implementation details
适用于病人我, their SCDA outcomey我is 1 if they experienced an SCDA event during the follow-up, and 0 otherwise.We adopted the balanced focal loss as the loss function54:$$L=-\sum _{i}{\alpha }_{i}{({\,y}_{i}-{p}_{i})}^{\gamma }\log {p}_{i}$$
(11)
在哪里
α我is a class-dependent scaling factor, andγis the focusing parameter that controls the level of how the model focuses on its mistakes and prioritizes improving on the hard examples, which was set asγ = 2 in this study.The LGE-CMR, EHR and CIR branch networks were first trained independently, and then MAARS was trained end-to-end with all the branch networks and the multimodal fusion module.All models were trained with a batch size of 64 and a maximum of 150 epochs with early stopping based on loss.
The Adam optimizer was used, withβ1 = 0.9,β2 = 0.999, and the learning rate was initially set at 1 × 10−3for the LGE-CMR branch network, 1 × 10−2for the EHR and CIR branch networks, and 3 × 10−2for the multimodal fusion and was adaptively adjusted during the training process.For the LGE-CMR branch network, the ViT haslViT = 8 transformer encoder blocks, eight heads for each attention module and dimensiond = 512.The EHR branch network used an FNN with two hidden layers and a latent dimension of 16. The CIR branch network used an FNN with one hidden layer and a latent dimension of 16. The encoded unimodal knowledge vectors have dimensionsξCMR ∈ r32,,,,ξEHR ∈ r16,,,,ξCIR ∈ r16。我们设定lMBTas 3 and the bottleneck fusion vector dimension as 8.Assessing model performance and clinical validation
Performance metrics
The values of metrics derived from the confusion matrix (BA, sensitivity and specificity) were computed at optimal probability decision thresholds selected to maximize Youden’s
jstatistic.When comparing the AI model’s performance to that of the clinical tools, we also adjusted the decision threshold by matching the sensitivities of the clinical tools to evaluate their specificities.All metrics were in the range of 0 to 1, with the baseline levels obtained by random chance being as follows: AUROC = 0.5, BA = 0.5, AUPRC = 0.03 and Bs = 0.25.
Internal and external validation
The internal model performance was assessed in a fivefold cross-validation of the JHH-HCM cohort on the patient level stratified by outcome.The training and test sets were split on the patient level;that is, all LGE-CMR scans corresponding to a given patient case were only present in either the training or validation set and never simultaneously partly in both.After five training folds, the model’s performance metrics were calculated based on the aggregation of all validation folds.
For the external performance evaluation, we trained the model using the entire JHH-HCM dataset (with 90% as the training set and 10% as the development set) and tested the model’s performance on the SHVI-HCM cohort.Of note, the model for external validation inherited the same hyperparameters as the internal model.
Model interpretability
We interpreted the MAARS network weights and predictions using attribution- and attention-based methods.
Shapley value
The EHR and CIR branch networks were interpreted using the Shapley value, which quantifies the incremental attribution of every input feature to the final prediction.The Shapley value32is based on the cooperative game theory and explains a prediction as a coalitional game played by the feature values.The Shapley value has a collection of desirable properties, including efficiency, symmetry, dummy and additivity.In this study, the Shapley values were estimated using a permutation formulation implemented in SHAP55。
Attention rollout
For the LGE-CMR branch network, we used a technique called attention rollout to quantify attention flows from the start to the end throughout the ViT.Formally, at transformer encoder blockl, the average of the attention matrices of all attention heads is一个l。The residual connection at each block is modeled by adding the identity matrix我to the attention matrix.Therefore, the attention rollout is recursively computed by
$$\begin{array}{c}{A}_{{\rm{Rollout}}}^{l}=\left({A}^{l}+I\right)\cdot{A}_{{\rm{Rollout}}}^{l-1}\end{array}$$
(12)
We explained the predictions of the LGE-CMR branch network using the attention rollout at the end of the ViT after flowing throughlViTtransformer blocks,\({A}_{{\rm{Rollout}}}^{{L}_{{\rm{ViT}}}}\)。统计分析这
p
values of clinical covariates between the internal and external cohorts were based on a two-sample Welch’st-test for numerical variables and the Mann–Whitney你test for categorical variables before data imputation.Kolmogorov–Smirnov tests for the risk score distributions were based on the aggregated predictions on all internal validation folds.The means and CIs of model performance metrics in the internal fivefold cross-validation were estimated using 200 bootstrapping samples of the aggregated predictions on all validation folds.The performance metrics in the external validation were calculated using model predictions on 200 bootstrapping resampled datasets of the SHVI-HCM cohort.The computations were based on the bias-corrected and accelerated bootstrap method.Pearson’srfor clinical covariates in the network interpretations was based on aggregated interpretations from all internal validation folds.Computational hardware and software
MAARS was built in Python 3.9 using packages including PyTorch 2.0, NumPy 1.23.5, Pandas 1.5.3, SciPy 1.10, scikit-learn 1.2.0, scikit-image 0.19.3, pydicom 2.3, SimpleITK 2.2.1 and SHAP 0.41.
Data preprocessing, model training and result analysis were performed on a machine with an AMD Ryzen Threadripper 1920X 12-core CPU and NVIDIA TITAN RTX GPUs, and on the Rockfish cluster at Johns Hopkins University using NVIDIA A100 GPU nodes, with NVIDIA software CUDA 11.7 and cuDNN 8.5.For a reference of the computational requirements of MAARS inference, on a machine with an AMD Ryzen 2700X 8-core CPU and an NVIDIA GeForce RTX 2060 GPU, the average processing time for inference is 0.034 s per patient using GPU or 0.086 s per patient using solely CPU.
报告摘要
有关研究设计的更多信息可在自然投资组合报告摘要链接到本文。
数据可用性
Patient data and individual-level source data in this article cannot be made publicly available without further consent and ethical approval owing to privacy concerns.The cardiac magnetic resonance imaging data, patient clinical data or source data can be provided by the authors pending approval from the Johns Hopkins University and Atrium Health institutional review boards and a completed material transfer agreement.Requests for these data should be sent to N.A.T.(ntrayanova@jhu.edu).Responses to requests are expected in a month.The final trained model weights are available athttps://gitlab.com/natalia-trayanova/maars-public。参考
Fishman, G. I. et al.
Sudden cardiac death prediction and prevention: report from a National Heart, Lung, and Blood Institute and Heart Rhythm Society Workshop.循环 122, 2335–2348 (2010).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Adabag, A. S., Luepker, R. V., Roger, V. L. & Gersh, B. J. Sudden cardiac death: epidemiology and risk factors.纳特。Cardiol牧师。 7, 216–225 (2010).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
John, R. M. et al.Ventricular arrhythmias and sudden cardiac death.柳叶刀 380, 1520–1529 (2012).
文章一个 PubMed一个 Google Scholar一个
Rea, T. D. & Page, R. L. Community approaches to improve resuscitation after out-of-hospital sudden cardiac arrest.循环 121, 1134–1140 (2010).
文章一个 PubMed一个 Google Scholar一个
Estes, N. A. M. 3rd Predicting and preventing sudden cardiac death.循环 124, 651–656 (2011).
文章一个 PubMed一个 Google Scholar一个
Merchant, F. M., Levy, W. C. & Kramer, D. B. Time to shock the system: moving beyond the current paradigm for primary prevention implantable cardioverter-defibrillator use.J. Am。Heart Assoc. 9, e015139 (2020).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Merchant, F. M., Jones, P., Wehrenberg, S., Lloyd, M. S. & Saxon, L. A. Incidence of defibrillator shocks after elective generator exchange following uneventful first battery life.J. Am。Heart Assoc. 3, e001289 (2014).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Semsarian, C., Ingles, J., Maron, M. S. & Maron, B. J. New perspectives on the prevalence of hypertrophic cardiomyopathy.J. Am。Coll.Cardiol。 65, 1249–1254 (2015).
文章一个 PubMed一个 Google Scholar一个
Maron, B. J. et al.Management of hypertrophic cardiomyopathy:JACCstate-of-the-art review.J. Am。Coll.Cardiol。 79, 390–414 (2022).
文章一个 PubMed一个 Google Scholar一个
Maron, B. J. et al.Prevalence of hypertrophic cardiomyopathy in a general population of young adults.Echocardiographic analysis of 4111 subjects in the CARDIA study.Coronary Artery Risk Development in (Young) Adults.循环 92, 785–789 (1995).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Ahluwalia, M., Kpodonu, J. & Agu, E. Risk stratification in hypertrophic cardiomyopathy: leveraging artificial intelligence to provide guidance in the future.JACC Adv. 2, 100562 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Börjesson, E., Svennblad, B., Wisten, A., Börjesson, M. & Stattin, E.-L.Symptoms and ECG changes precede sudden cardiac death in hypertrophic cardiomyopathy—a nationwide study among the young in Sweden.PLoS ONE 17, e0273567 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Calò, L. et al.The value of the 12-lead electrocardiogram in the prediction of sudden cardiac death.欧元。Heart J. Suppl. 25, C218–C226 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
van Velzen, H. G. et al.Value of genetic testing for the prediction of long-term outcome in patients with hypertrophic cardiomyopathy.是。J. Cardiol. 118, 881–887 (2016).
文章一个 PubMed一个 Google Scholar一个
Hernesniemi, J. A. et al.Predicting sudden cardiac death using common genetic risk variants for coronary artery disease.欧元。心脏。 36, 1669–1675 (2015).
文章一个 PubMed一个 Google Scholar一个
Guo, L. et al.Genetic variants associated with unexplained sudden cardiac death in adult white and African American individuals.贾马心脏。 6, 1013–1022 (2021).
文章一个 PubMed一个 Google Scholar一个
Augusto, J. B. et al.Diagnosis and risk stratification in hypertrophic cardiomyopathy using machine learning wall thickness measurement: a comparison with human test–retest performance.柳叶刀数字。健康 3, e20–e28 (2021).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Navidi, Z. et al.Interpretable machine learning for automated left ventricular scar quantification in hypertrophic cardiomyopathy patients.PLOS数字。健康 2, e0000159 (2023).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Laubenbacher, R., Mehrad, B., Shmulevich, I. & Trayanova, N. Digital twins in medicine.纳特。计算。科学。 4, 184–191 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
O’Hara, R. P. et al.Personalized computational heart models with T1-mapped fibrotic remodeling predict sudden death risk in patients with hypertrophic cardiomyopathy.Elife 11, e73325 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Kochav, S. M. et al.Predicting the development of adverse cardiac events in patients with hypertrophic cardiomyopathy using machine learning.int。J. Cardiol. 327, 117–124 (2021).
文章一个 PubMed一个 Google Scholar一个
Smole, T. et al.A machine learning-based risk stratification model for ventricular tachycardia and heart failure in hypertrophic cardiomyopathy.计算。生物。医学 135, 104648 (2021).
文章一个 PubMed一个 Google Scholar一个
Bhattacharya, M. et al.Identifying ventricular arrhythmias and their predictors by applying machine learning methods to electronic health records in patients with hypertrophic cardiomyopathy (HCM-VAr-Risk Model).是。J. Cardiol. 123, 1681–1689 (2019).
文章一个 PubMed一个 Google Scholar一个
Zhao, K. et al.Machine learning in hypertrophic cardiomyopathy: nonlinear model from clinical and CMR features predicting cardiovascular events.JACC Cardiovasc.成像 17, 880–893 (2024).
文章一个 PubMed一个 Google Scholar一个
Kolk, M. Z. H. et al.Multimodal explainable artificial intelligence identifies patients with non-ischaemic cardiomyopathy at risk of lethal ventricular arrhythmias.科学。代表。 14, 14889 (2024).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ommen, S. R. et al.2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines.循环 142, e558–e631 (2020).
PubMed一个 Google Scholar一个
Arbelo, E. et al.2023 ESC guidelines for the management of cardiomyopathies: developed by the task force on the management of cardiomyopathies of the European Society of Cardiology (ESC).欧元。心脏。 44, 3503–3626 (2023).
文章一个 CAS一个 PubMed一个 Google Scholar一个
O’Mahony, C. et al.A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM Risk-SCD).欧元。心脏。 35, 2010–2020 (2014).
文章一个 PubMed一个 Google Scholar一个
Dosovitskiy, A. et al.An image is worth 16×16 words: transformers for image recognition at scale.在International Conference on Learning Representations 2021 https://openreview.net/pdf?id=YicbFdNTTy(2021)。
Nagrani, A. et al.Attention bottlenecks for multimodal fusion.在35th Conference on Neural Information Processing Systems (NeurIPS 2021) https://proceedings.neurips.cc/paper_files/paper/2021/file/76ba9f564ebbc35b1014ac498fafadd0-Paper.pdf(NeurIPS, 2021).
Corianò, M. et al.Deep learning-based prediction of major arrhythmic events in dilated cardiomyopathy: a proof of concept study.PLoS ONE 19, e0297793 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Shapley, L. A value forn-person games.Contributions to the Theory of Games II(1953) 307–317.在Classics in Game Theory(ed. Kuhn, H. W.) 69–79 (Princeton Univ. Press, 1997);https://doi.org/10.1515/9781400829156-012
O’Mahony, C. et al.Effectiveness of the 2014 European Society of Cardiology guideline on sudden cardiac death in hypertrophic cardiomyopathy: a systematic review and meta-analysis.心 https://doi.org/10.1136/heartjnl-2018-313700(2018)。
文章一个 PubMed一个 Google Scholar一个
Dong, Y. et al.Validation of the 2020 AHA/ACC risk stratification for sudden cardiac death in Chinese patients with hypertrophic cardiomyopathy.正面。心脏。医学 8, 691653 (2021).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zegkos, T. et al.Validation of the new American College of Cardiology/American Heart Association Guidelines for the risk stratification of sudden cardiac death in a large Mediterranean cohort with hypertrophic cardiomyopathy.Hellenic J. Cardiol. 63, 15–21 (2022).
文章一个 PubMed一个 Google Scholar一个
O’Mahony, C. et al.International external validation study of the 2014 European Society of Cardiology Guidelines on Sudden Cardiac Death Prevention in Hypertrophic Cardiomyopathy (EVIDENCE-HCM).循环 137, 1015–1023 (2018).
文章一个 PubMed一个 Google Scholar一个
Vriesendorp, P. A. et al.Validation of the 2014 European Society of Cardiology guidelines risk prediction model for the primary prevention of sudden cardiac death in hypertrophic cardiomyopathy.循环。Arrhythm.Electrophysiol. 8, 829–835 (2015).
文章一个 PubMed一个 Google Scholar一个
Wang,J。等。Radiomics analysis derived from LGE-MRI predict sudden cardiac death in participants with hypertrophic cardiomyopathy.正面。心脏。医学 8, 766287 (2021).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Popescu, D. M. et al.Arrhythmic sudden death survival prediction using deep learning analysis of scarring in the heart.纳特。心脏。res。 1, 334–343 (2022).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical AI.纳特。医学 28, 1773–1784 (2022).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Liang, P. P., Zadeh, A. & Morency, L.-P.Foundations and trends in multimodal machine learning: principles, challenges, and open questions.ACM Comput.幸存。 https://doi.org/10.1145/3656580(2024)。
Maron, M. S. et al.Effect of left ventricular outflow tract obstruction on clinical outcome in hypertrophic cardiomyopathy.N. Engl.J. Med。 348, 295–303 (2003).
文章一个 PubMed一个 Google Scholar一个
Lu, D. et al.Clinical outcomes in patients with nonobstructive, labile, and obstructive hypertrophic cardiomyopathy.J. Am。Heart Assoc. 7, e006657 (2018).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Parcharidou, D. et al.The natural history of hypertrophic cardiomyopathy in a large Mediterranean cohort.J. Cardiovasc.医学(Hagerstown) 21, 428–434 (2020).
文章一个 PubMed一个 Google Scholar一个
Pozios, I. et al.Rest and stress longitudinal systolic left ventricular mechanics in hypertrophic cardiomyopathy: implications for prognostication.J. Am。Soc。Echocardiogr. 31, 578–586 (2018).
文章一个 PubMed一个 Google Scholar一个
Maron, M. S. et al.Contemporary natural history and management of nonobstructive hypertrophic cardiomyopathy.J. Am。Coll.Cardiol。 67, 1399–1409 (2016).
文章一个 PubMed一个 Google Scholar一个
Pozios, I. et al.Comparison of outcomes in patients with nonobstructive, labile-obstructive, and chronically obstructive hypertrophic cardiomyopathy.是。J. Cardiol. 116, 938–944 (2015).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Riley, R. D. et al.Evaluation of clinical prediction models (part 3): calculating the sample size required for an external validation study.BMJ 384, e074821 (2024).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Schinkel, A. F. L. et al.Outcome and complications after implantable cardioverter defibrillator therapy in hypertrophic cardiomyopathy: systematic review and meta-analysis.循环。Heart Fail. 5, 552–559 (2012).
文章一个 CAS一个 PubMed一个 Google Scholar一个
Fahmy, A. S. et al.Radiomics of late gadolinium enhancement reveals prognostic value of myocardial scar heterogeneity in hypertrophic cardiomyopathy.JACC Cardiovasc.成像 17, 16–27 (2024).
文章一个 PubMed一个 Google Scholar一个
Bhattacharya, M. et al.Machine learning methods for identifying atrial fibrillation cases and their predictors in patients with hypertrophic cardiomyopathy: the HCM-AF-Risk Model.CJC Open 3, 801–813 (2021).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Popescu, D. M. et al.Anatomically informed deep learning on contrast-enhanced cardiac magnetic resonance imaging for scar segmentation and clinical feature extraction.心脏。数字。Health J. https://doi.org/10.1016/j.cvdhj.2021.11.007(2021)。
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
van Buuren, S. & Groothuis-Oudshoorn, K. MICE: multivariate imputation by chained equations in R.J. Stat.Softw. 45, 1–67 (2011).
文章一个 Google Scholar一个
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection.IEEE Trans。模式肛门。马赫。Intell。https://doi.org/10.1109/TPAMI.2018.2858826(2020)。Lundberg, S. M. & Lee, S.-I.
A unified approach to interpreting model predictions.在31st Conference on Neural Information Processing Systems (NIPS 2017) https://proceedings.neurips.cc/paper_files/paper/2017/file/8a20a8621978632d76c43dfd28b67767-Paper.pdf(2017)。
致谢
This work is supported by National Institutes of Health grants R01HL166759 (N.A.T.), R01HL174440 (N.A.T.) and R35HL1431598 (A.G.H.) and a Leducq Foundation grant (N.A.T.).
竞争利益
作者没有宣称没有竞争利益。
同行评审
同行评审信息
自然心血管研究
thanks Declan O’Regan and Jagmeet P. Singh for their contribution to the peer review of this work.附加信息
Publisher’s note
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。扩展数据
Extended Data Fig. 1 Personalized interpretation of EHR branch network predictions.
Shown are Shapley value-based interpretations for a patient who did not experience SCDA (top panel) and a patient who experienced SCDA (bottom panel).
The plots are read from bottom up: the number under the x-axis is the mean risk score for the cohort (0.389);the blue and red bars are the Shapley values associated with the input covariates, representing the effect of each covariate on the output risk score;finally, with the effects from all the covariates, the model reaches its final prediction on top of the plot (value of f(x)).
Extended Data Fig. 2 Personalized interpretation of CIR branch network.
Shown are Shapley value-based interpretations for a patient who did not experience SCDA (top panel) and a patient who experienced SCDA (bottom panel).The plots are read from bottom up: the number under the x-axis is the mean risk score for the cohort (0.545);the blue and red bars are the Shapley values associated with the input covariates, representing the effect of each covariate on the output risk score;finally, with the effects from all the covariates, the model reaches its final prediction on top of the plot (value of f(x)).
Extended Data Fig. 3 Study flowcharts.
The flowchart (一个) shows the eligibility assessment of patients from the JHH-HCM cohort, a subset of which was used for cross-validation.The flowchart (b) shows the eligibility assessment of patients from the SHVI-HCM cohort for external validation.Extended Data Table 1 List of EHR covariates used in MAARS
权利和权限
开放访问
This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material.您没有根据本许可证的许可来共享本文或部分内容的改编材料。The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.要查看此许可证的副本,请访问http://creativecommons.org/licenses/by-nc-nd/4.0/。重印和权限
引用本文
Lai, C., Yin, M., Kholmovski, E.G.
等。Multimodal AI to forecast arrhythmic death in hypertrophic cardiomyopathy.Nat Cardiovasc Res(2025)。https://doi.org/10.1038/s44161-025-00679-1
已收到:
公认:
出版:
doi:https://doi.org/10.1038/s44161-025-00679-1