Detecting structural heart disease from electrocardiograms using AI
作者:Elias, Pierre
Main
Structural heart disease (SHD) is a growing epidemic that remains substantially underdiagnosed. SHD encompasses pathologies that affect the valves, walls or chambers of the heart, including valvular heart disease (VHD), right- and left-sided heart failure, pulmonary hypertension and left ventricular hypertrophy6. SHD represents more than US $100 billion in annual direct and indirect costs in the USA, a number that will continue to rise as the disease burden increases7,8,9. The impact of these diseases is profound, with heart failure and VHD affecting an estimated 64 million and 75 million people, respectively, with prevalence increasing10,11,12,13. Despite its clinical importance, SHD remains underdiagnosed. A study of 2,500 people 65 years of age or older found that 4.9% were previously diagnosed with clinically significant (graded moderate or severe) VHD and found another 6.4% with undiagnosed VHD more than doubling the overall prevalence14. Finding patients with SHD earlier in the disease process has been shown to reduce mortality, decrease costs and improve quality of life, but getting to diagnosis remains challenging1,15,16. For at least two forms of SHD, heart failure and VHD, symptoms can be attributed to many potential diagnoses and are often only present late in the disease course. All forms of SHD can be definitively diagnosed with echocardiography, but cost, required expertise and appropriate patient selection limit its total use. Thus, there remains a critical need to better risk stratify patients and determine who should be referred for echocardiography to improve rates of SHD diagnosis and early treatment.
The application of deep learning, a subset of artificial intelligence (AI), has been shown to be beneficial in the detection of specific heart diseases from the 12-lead electrocardiogram (ECG). This includes conditions such as aortic stenosis, low left ventricular ejection fraction (LVEF) and low left ventricular hypertrophy, as well as composites of many valvular diseases3,17,18,19,20,21. More general AI-ECG models have also been developed that can accurately detect a composite of low left ventricular systolic dysfunction, low left ventricular hypertrophy and moderate or greater VHD4. As model precision (positive predictive value) is affected by outcome prevalence, the use of a composite prediction target takes advantage of increased outcome prevalence (summation of component prevalence, if independent) to achieve a higher model precision than possible with models trained for any individual components. Moreover, when the disease label components share a clinical diagnostic pathway (for example, requiring confirmation with echocardiography), these precision gains are achievable with no further operational cost, as a high-risk score is suggestive for echocardiography referral in all cases. Challenges with these models include ensuring model performance across a wide range of disease states, clinical contexts and patient demographics. Most importantly, these models and the underlying training data are typically proprietary, limiting comparisons and broader evaluation of generalizability.
This study was designed to use data from a large and diverse hospital system to achieve the following aims: (1) develop a deep learning ECG model that can accurately detect a broad array of SHDs; (2) assess the generalizability of model performance across institutions, patient demographics and clinical contexts; (3) test these technologies in a pilot clinical trial to determine whether they can be used to practicably detect undiagnosed heart disease; and (4) publicly release both an SHD detection model and a large de-identified, ECG dataset with curated echocardiography-derived labels to spur further research.
Model development and validation
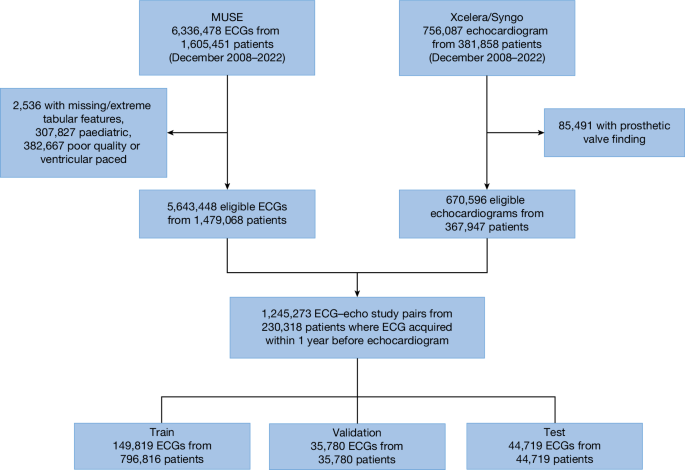
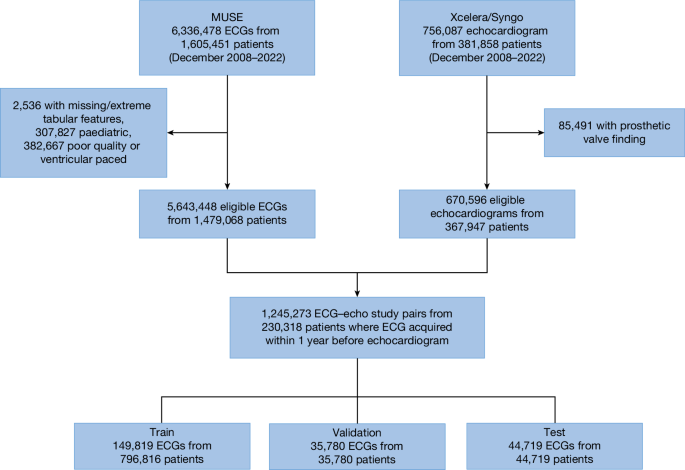
We curated a dataset comprising 1,245,273 ECG–echocardiogram pairs from 230,318 unique patients (aged 18 years or above) collected between December 2008 and 2022 at one of eight New York–Presbyterian (NYP) affiliated hospitals (Fig. 1). This dataset was designated as the NYP multicentre cohort. The data were split at a patient level into train (149,819 unique patients with 796,816 ECG–echocardiogram pairs), validation (35,780 unique patients with 35,780 ECG–echocardiogram pairs) and test (44,719 unique patients with 44,719 ECG–echocardiogram pairs) sets with patient characteristics described in Table 1. Of note, only the most recent ECG–echocardiogram pair per patient was retained for validation and testing.
The deep learning model was trained and tested using data from an eight-hospital system (NYP Hospital). ECG data were accessed using the MUSE system with removal of ECGs with missing age, sex and patient identifier, poor study quality designation by machine recommending repeating of ECG or presence of ventricular pacing. Echocardiogram data were accessed using hospital systems with removal of patients with repaired or replaced heart failures. This yielded 1.2 million ECG–echocardiogram pairs in 230,018 unique patients with data split into train, validation and test sets.
The presence of SHD was a composite of the following conditions identified on clinical echocardiography reports on the basis of appropriate echocardiography guidelines: low LVEF less than or equal to 45%; maximum low left ventricular wall thickness greater than or equal to 1.3 cm; moderate or severe right ventricular dysfunction; pulmonary hypertension (pulmonary artery systolic pressure (PASP) greater than or equal to 45 mm Hg or tricuspid regurgitation jet velocity greater than or equal to 3.2 m s−1); moderate or severe aortic stenosis, aortic regurgitation, mitral regurgitation, tricuspid regurgitation or pulmonary regurgitation, or a moderate or large pericardial effusion22,23,24,25. These cutoffs were chosen to generally correlate with clinically accepted definitions for moderate or greater pathology and to identify patients with low left ventricular systolic dysfunction that would be eligible for guideline-directed medical therapy as determined in recent studies26,27,28,29,30. For an ECG to be labelled as being ‘positive’ for a disease, it must have been performed within 1 year before an echocardiogram with SHD. In patients without SHD (confirmed by at least one ‘negative’ echocardiogram), all ECGs before the most recent echo were labelled as negative and included in the study. Using these definitions, the prevalence of SHD in the test set was 36%.
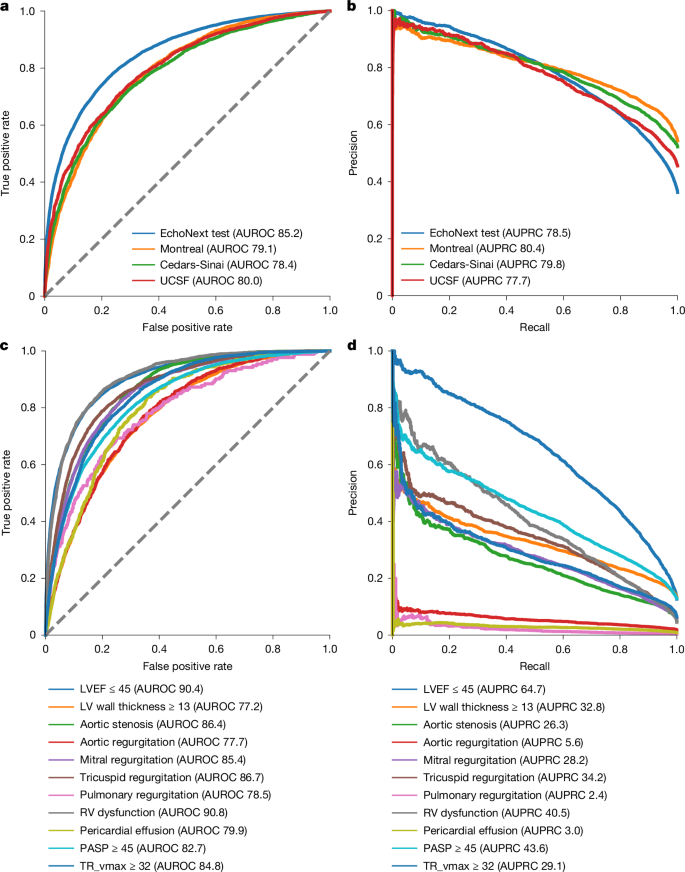
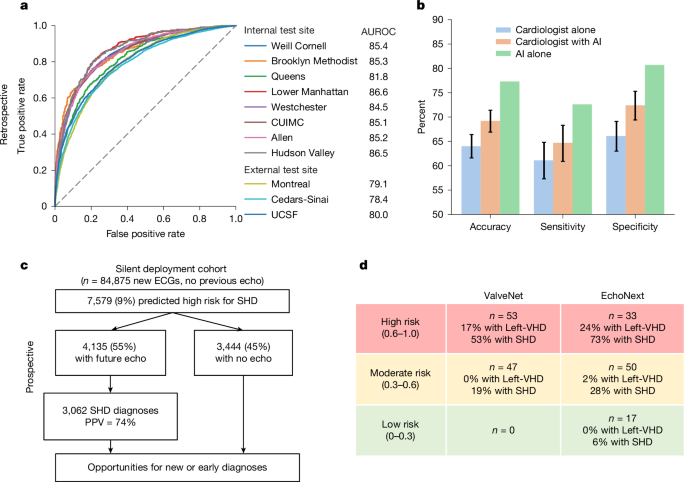
These data were used to train EchoNext, a convolutional neural network model (Supplementary Table 1), to predict the presence of SHD using ECG traces and seven standard values included on ECGs (age; sex (captured from the ECG demographic data); atrial rate; ventricular rate; pulmonary regurgitation interval; Q wave, R wave and S wave (QRS) duration; and corrected Q wave-to-T wave interval). EchoNext performance on the NYP multicentre test set was high and well calibrated (Figs. 2 and 3 and Supplementary Fig. 1), with an area under the receiver operating characteristic curve (AUROC) of 85.2% (95% confidence interval (CI) 84.5–85.9%), area under the precision–recall curve (AUPRC) of 78.5% (95% CI 77.2–79.6%) and diagnostic odds ratio of 12.8 (95% CI 11.6–14.1) (Fig. 2). In addition to the SHD composite label, EchoNext was trained as a multitask classifier to also predict each individual disease label within the composite, to better assess label collinearity and ensure consistent predictions among highly correlated labels (Supplementary Fig. 4). For example, pulmonary regurgitation is highly correlated with tricuspid regurgitation max velocity and right ventricular dysfunction. Model performance varied across each of these individual components with right ventricular (AUROC 91%) and low left ventricular systolic dysfunction (90%) as the best performing (Fig. 2 and Supplementary Table 2). The lowest performance was seen for low left ventricular wall thickness (AUROC 77%), AR (78%), pulmonary regurgitation (79%) and pericardial effusion (80%). We further evaluated model performance across NYP hospitals, clinical contexts and with respect to patient age and race and/or ethnicity (Table 2). Across hospitals (AUROC range 82–87%) (Fig. 3) and clinical contexts (AUROC range 79–84%), the model showed stable, generalizable performance. Similarly, there were no clinically relevant differences in model performance by race and/or ethnicity or sex; model discrimination was slightly improved in younger populations, consistent with patterns reported in other analyses3,17,20,31.
Performance of the model in detection of individual and compositive SHDs. a,b, By AUROC (a) and AUPRC (b), the model had high performance in detection of SHD in the internal eight-hospital NYP system test set and three geographically distinct external test sets (Montreal Heart, Cedars-Sinai and University of California San Francisco (UCSF)). c, Individual disease models had the highest performance in the detection of reduced low left ventricular (LV) and right ventricular (RV) systolic function by AUROC with favourable performance for other disease states. d, Assessment of the AUPRC for the individual disease states is highly dependent on the underlying prevalence of the individual disease states. TR, tricuspid regurgitation. Dashed lines (a,c) indicate random classifier.
a, In test sets of held-out patients at these sites, as well as in three geographically distinct external test sets, the model demonstrated high accuracy, as demonstrated by AUROC. Dashed line indicates random classifier. b, In a survey of ECGs shown to cardiologists to assess for the presence of SHD, the AI model demonstrated superior performance in SHD detection compared with cardiologists alone or cardiologists given the EchoNext risk score (n = 3,200 cardiologist interpretations). Error bars show the CIs derived from results across 13 cardiologists; because the AI model was run once on the entire set of 150 ECGs, there are no error bars for the ‘AI alone’ results. c, This model was evaluated in a temporally distinct held-out set with similar accuracy, with 45% (n = 3,444) of patients labelled as high risk by the model failing to undergo echocardiography as part of routine clinical care. d, The clinical use of AI-ECG to detect SHD was evaluated in a single-arm, single-site, open-label pilot clinical trial, DISCOVERY, with stratified recruitment of patients (N = 100) with an ECG but no previous echocardiogram. This trial used a related ECG model, ValveNet, which was trained to detect left-sided VHD (Left-VHD) with patients selected stratified sampling by their AI-ECG scores. This trial showed a high-level of discrimination in the detection of Left-VHD (primary endpoint) and SHD (secondary endpoint), and post hoc assessment using the second-generation EchoNext model demonstrated an even greater degree of risk stratification with 73% of patients in the highest risk and 6% of patients in the lowest-risk groups being found to have SHD. Left-VHD, moderate or severe aortic stenosis, aortic regurgitation or mitral regurgitation; SHD, LVEF less than or equal to 45%, low left ventricular wall thickness greater than or equal to 1.3 cm, moderate or severe right ventricular dysfunction, any moderate or severe VHD, PASP greater than or equal to 45 mm Hg or a moderate or large pericardial effusion.
Finally, versions of the model alternatively trained on more restrictive phenotype definitions (‘severe’ SHD; Supplementary Information, Supplementary Fig. 2 and Supplementary Table 3) or using different partitions of the NYP multicentre cohort (Supplementary Information, Supplementary Fig. 3 and Supplementary Tables 4–8) showed minimal differences in performance. When the model was trained on data from four of the eight NYP hospitals and tested on independent data from the four unseen hospitals, the performance change was minimal. The same was true when switching the hospitals used for training and testing. Furthermore, performance was stable for both academic and community hospitals. Performance remained robust when tested on different combinations of component disease labels (left or right ventricular dysfunction, all valvular disease, left-sided heart disease and right-sided heart disease).
External validation
The performance of the model was tested in three external cohorts from Cedars-Sinai Medical Center (n = 10,177 patients), the Montreal Heart Institute (n = 10,862) and the University of California San Francisco Medical Center (n = 6,106). SHD prevalence was higher in the external sites (54%, 52% and 46%, respectively) compared with SHD prevalence in the NYP cohort (36%). EchoNext showed a 5–7% drop in AUROC (78–80%) in these external cohorts as compared to individual hospitals within the NYP multicentre cohort (Fig. 3). At a fixed sensitivity of 70%, the external cohorts showed comparable positive predictive value, but a 10% drop in specificity (Supplementary Table 10). These differences may be attributed to the large differences in disease prevalence and other patient demographic features compared with the model training population (Supplementary Table 9). Similar or greater performance variation has been seen in the validation of non-AI detection technologies such as Troponin assays and screening mammography.
Silent deployment validation
We further sought to evaluate model performance in a temporally distinct NYP patient cohort, which was more reflective of the real-world intended-use population for this model. On the 124,027 ECGs acquired between 1 January 2023 and 16 September 2023 from 84,875 unique patients with no previous echocardiogram, EchoNext was automatically run in the background and the model prediction was stored for future use. None of these patients were included in the original training, validation or test datasets. Patients were then monitored to see if they received an echocardiogram at any of the eight hospitals at a future date. In this cohort, 18% (15,094 patients) underwent their first echocardiogram as part of routine clinical care after the ECG; of which, 38% (5,744 patients) were newly diagnosed with SHD. In this subset with echocardiography follow-up, the model again generalized well, with similar performance (AUROC 83%, AUPRC 81%) to the retrospective development cohort. Using the predefined model score cutoff of 0.6, 27% (4,135 patients) of the ECG and/or echocardiogram population were predicted as high risk, corresponding to a precision (positive predictive value) and recall (sensitivity) of 74% and 53%, respectively.
Among the 69,781 patients with an ECG but no follow-up echocardiogram, 3,444 (5%) were predicted as high risk for SHD at the same cutoff as above. Given that the actual prevalence of disease in this population is unknown, Table 3 provides estimates of model precision as a function of varying prevalence and chosen sensitivity. For example, if SHD prevalence is 10% in this population, the projected positive predictive value would be 46.5% at 50% sensitivity. Thus, out of the 3,444 patients at high-risk, an estimated 1,998 patients could have newly diagnosed SHD through model-directed intervention.
Model performance versus cardiologists
We created a custom survey to test the ability of cardiologists to detect SHD using ECGs compared with the EchoNext model. A set of 150 ECGs was abstracted from the NYP multicentre test set with a similar SHD prevalence (41%) and age distribution (mean 67.0 ± 19.6) as the entire dataset. ECGs were de-identified and built into a custom interface for the survey. For non-AI-assisted reviews, the cardiologist was presented with the ECG waveform, standard ECG-derived features (atrial or ventricular rate, pulmonary regurgitation interval, QRS duration, corrected Q wave-to-T wave interval), age and sex of the patient and asked to state whether they thought the patient had SHD. Reviews were completed in blocks of 50 ECGs such that, after completing a block of non-AI-assisted reviews, the same set of 50 ECGs was repeated with the EchoNext score added to the survey interface (AI-assisted reviews). Each cardiologist could complete up to 300 reviews total (150 non-AI-assisted results and 150 AI-assisted results).
A total of 13 cardiologists completed 3,200 ECG survey reviews (1,600 without and 1,600 with AI assistance, average 246 ECGs per cardiologist; Supplementary Table 11). In the set of 150 ECGs, the EchoNext model had an accuracy of 77.3%, sensitivity 72.6% and specificity of 80.7%. For the 1,600 non-AI-assisted reviews, the accuracy of the cardiologists was 64.0% (95% CI 61.6–66.4%) with sensitivity 61.1% (95% CI 57.3–64.8%) and specificity 66.1% (95% CI 63.0–69.1%). Notably, the accuracy of the cardiologists differed between clinically normal and abnormal ECGs (69% versus 62%, respectively), whereas EchoNext performed equally well (77% for both) despite the substantial difference in SHD prevalence (25% versus 46.5% in normal versus abnormal ECGs) (Supplementary Table 12). With AI assistance, the accuracy of the cardiologists significantly improved to 69.2% (95% CI 66.9–71.4%) with sensitivity 64.7% (95% CI 60.9–68.3%) and specificity 72.4% (95% CI 69.4–75.3%). Thus, whereas AI assistance for cardiologists in this task improved predictive accuracy, the combined performance still lagged that of the AI alone. This task focused on using the ECG for SHD detection without taking advantage of other information (clinical history, physical exam, other testing data) that would typically be available to a physician in clinical practice.
Prospective validation
Before development of EchoNext, study investigators had created ValveNet, a similarly architected AI-ECG model trained to detect moderate or greater left-sided VHD (specifically aortic stenosis, aortic regurgitation and mitral regurgitation), a subset of SHD3. To test the model’s ability to detect clinically significant cardiac disease, we designed the DISCOVERY (detecting SHD using deep learning on an electrocardiographic waveform array) trial, which was a 100-patient open-label stratified sampling prospective trial recruiting patients on the basis of their ValveNet risk score. Adult patients were eligible if they had a digital 12-lead ECG performed at Columbia University and had no history of an echocardiogram within the last 3 years in our system, no history of left-sided VHD and no dementia or other non-cardiac life-limiting disease with expected survival less than 1 year. Eligible patients were recruited by their ValveNet score, which was divided into prespecified tertiles of risk (0–0.3, 0.3–0.6, greater than 0.6). The lowest-risk group was excluded due to a very low predicted risk of cardiac disease. Consented patients underwent an echocardiogram. The primary endpoint was detection of moderate or severe aortic stenosis, aortic regurgitation or mitral regurgitation. The key secondary endpoint was detection of all forms of clinically significant SHD using the same definition and thresholds as EchoNext. Critically important findings were communicated with patients and physicians, and appropriate clinical follow-up was coordinated by study investigators for newly diagnosed disease.
The median age of recruited patients was 80 years (interquartile range 72–86) and 43% were male (Supplementary Table 16). A total of 53 patients with high-risk ValveNet scores were recruited, with 17% positive for moderate or greater left-sided VHD and 53% positive for SHD. A total of 47 patients were recruited with moderate-risk ValveNet scores, with 0% positive for left-sided VHD and 19% positive for SHD. There was a significant difference in the number of patients positive for left-sided VHD (P = 0.005) and SHD (P = 0.003) when comparing high- to moderate-risk ValveNet scores.
After trial completion the 100 patients’ ECGs were retrospectively analysed using EchoNext and stratified into high-, moderate- and low-risk groups. The rates of disease were strongly correlated within these groups as follows: high risk (n = 33, 24% with left-sided VHD and 73% with SHD), moderate risk (n = 50, 2% with left-sided VHD, 28% with SHD) and low risk (n = 17, 0% with left-sided VHD and 6% with SHD). All differences between risk groups for EchoNext were significant (P = 0.002 for left-sided VHD and P < 0.001 for SHD). Individual disease outcomes stratified by risk groups are summarized in Supplementary Table 17.
AI benchmark in SHD
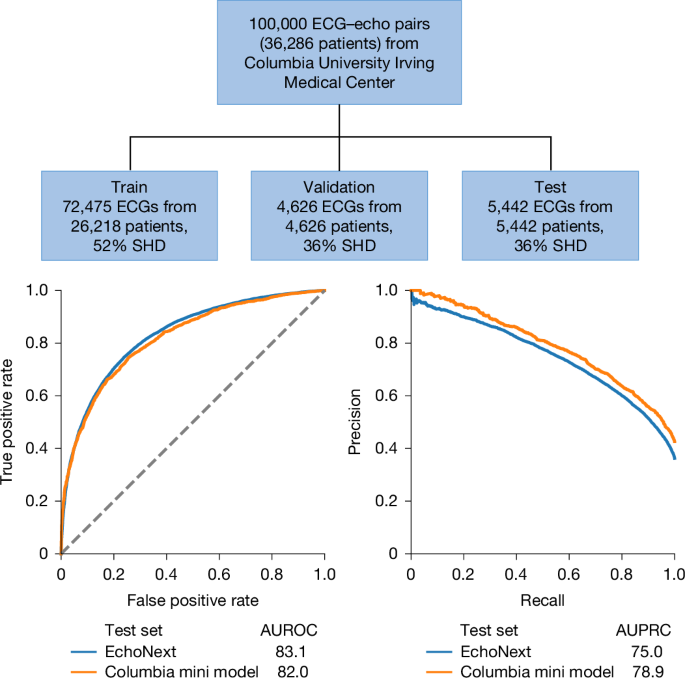
To facilitate future research in this space and to create publicly available data for model benchmarking, we are releasing, with publication of this paper, a de-identified and annotated set of ECG data. These data comprise 100,000 ECGs from 36,286 unique patients from Columbia University Irving Medical Center (Fig. 4). These ECGs represent a subset of the NYP multicentre Cohort and were labelled with respect to SHD status (as well as the individual components) on the basis of matched echocardiograms following the same procedure. We divided the ECGs into train, validation and test sets and trained a de novo model within this population, hereafter referred to as the Columbia mini-model. The SHD prevalence in this test set was 43%.
A 100,000 ECG dataset is being released from data from Columbia University Irving Medical Center as part of this paper. These data consist of the ECG waveform, ECG tabular features and paired echocardiographic data. A model trained and tested on this dataset demonstrated similar performance in SHD detection as the multisite model that served as the primary analysis in this study when assessed by AUROC and AUPRC. Dashed line indicates random classifier.
The Columbia mini-model was highly performant for SHD detection across all eight hospitals, with AUROC of 82.0% (95% CI 80.9–83.0%) (Fig. 4). Of note, despite the smaller and single-centre training set, this model’s performance was only modestly lower than the full multicentre-trained EchoNext model, with AUROC in the same multicentre test set of 83.1%.
This dataset, including the ECG waveform, demographics and ECG-specific tabular information, as well as all the corresponding echocardiographic labels, was de-identified according to standard practices. All date information was shifted randomly on a per patient level by more than 1 year preserving time elapsed between individual studies for the same patient. To spur further research and serve as an ECG benchmark, this dataset, as well as the preprocessing code and Columbia mini-model weights, are available at the EchoNext Library with further instructions for use available in the Supplementary Information. It will also be expanded on with further studies and modalities over time.
Discussion
We present here the development and performance of the EchoNext ECG deep learning model for the detection of SHD, and a pilot prospective trial of an AI-ECG model to detect SHD3,4.The principal findings of this study are as follows: (1) the EchoNext model accurately detects a broad composite of clinically relevant SHDs that would warrant ordering an echocardiogram, which was strengthened by the multilabel approach to best capture collinearity and correlation across component disease labels; (2) EchoNext generalized across 11 hospitals from 4 health systems (both inclusive and exclusive of the model development cohort); (3) performance was robust with respect to patient demographics (age, sex, race and/or ethnicity) across clinical contexts in a highly diverse patient population; (4) EchoNext had superior accuracy, sensitivity and specificity in detecting SHD from ECGs compared to cardiologists both when given the AI prediction to consult and when not; and (5) the ability of AI-ECG analysis to prospectively detect undiagnosed cardiac disease with sufficient positive predictive value was confirmed in the DISCOVERY trial. Moreover, the release of data, code and model weights from this work can serve as a benchmark and catalyst for future research in this field.
Expanding ECG use through AI
The ECG remains a core diagnostic test in cardiology even as we reach the 100-year anniversary of Dr Einthoven’s receipt of the 1924 Nobel Prize in Physiology and Medicine. The history of the ECG has been one of relentless technological progress, beginning with a 600-pound galvanometer electrocardiograph only available in research settings, to digital ECGs that can be performed anywhere and are available everywhere, including at home and on the wrist32,33. The work over the past decade applying deep learning methodologies to the ECG continues this trend, opening new insights and approaches for heart disease detection.
In this work, we present direct evidence for some of this new use through the comparison of the EchoNext AI model to board-certified cardiologists’ performance in detecting SHD from ECGs. To be clear, explicit detection of most SHD from ECG—particularly without other aspects of clinical history and physical exam—is not a standard clinical practice and therefore, as expected, the cardiologists were only modestly successful in the task. By comparison, the performance of the EchoNext model was significantly superior, improving on both diagnostic sensitivity and specificity compared with the human experts. Together, these data demonstrate the potential for AI to help further expand the clinical and diagnostic use for an already broadly used and broadly accessible test. The fact that EchoNext alone performed significantly better than cardiologists even when given the AI results warrants further exploration. Clinical experts may still lack trust in AI systems, particularly in situations in which performance achieved by AI was not previously thought possible. For example, cardiologists do not rely on ECG findings to determine whether a patient is likely to have a reduced LVEF, but reduced LVEF is one of the highest performing component disease predictions of EchoNext (AUROC 90.4) (Fig. 2c). Further exploration is needed to identify optimal strategies that integrate the specialized ECG interpretation from AI with the clinician’s more extensive, diverse knowledge to improve detection of patients with subtle clinical signs or those who may lack consistent clinical care.
Medical AI models must advance from interpreting one study at one point in time to making a comprehensive patient-level prediction. Future models may benefit from incorporating multimodality (for example, integrating chest X-rays, laboratory results and ECGs) and multitemporality (for example, using all past ECGs of a patient). These advancements aim to create a comprehensive prediction of a patient’s risk. However, this approach presents several challenges. As data requirements increase, so does the risk of confounding and label leakage, potentially leading to models that appear to perform well but generalize poorly. Furthermore, the complexity of integration and adoption of such models increases considerably.
Translation to clinical care
Bridging the gap between retrospective development of clinical AI models and studying their efficacy in improving clinical care is paramount. There have been few prospective studies of AI-ECG models thus far. The DISCOVERY trial is the first focused specifically on the detection of VHD and the broader composite of all SHD. The positive findings from this trial provided critical confirmation that (1) application of an AI-ECG model to a real-world intended-use population (that is, a population with lower disease prevalence than the development cohort) still yielded clinically meaningful performance (in this case, a positive predictive value of greater than 50% for SHD); and (2) beyond simple binary risk prediction, the burden of observed disease varied across levels of model-predicted risk (moderate versus high), potentially enabling performance tuning for different use cases.
In light of these positive trial results, other aspects of this AI-ECG model lend themselves well to translation into clinical practice. First, the intended clinical action to be taken on the basis of model output is clear: a high-risk result should be considered for an echocardiogram. The fact that an abnormal ECG is already a common indication for echocardiography further reduces the barrier for this response. The use of a broad composite outcome, defined as all causes of clinically significant SHD, as a model target is a deliberate strategy to optimize the positive predictive value of the model. Whereas the truly ‘optimal’ positive predictive value from the varied perspectives of providers, patients and payors is yet to be defined, the fact that EchoNext further improved risk stratification for SHD within the trial cohort compared to ValveNet supports this motivation. Further prospective studies testing the next-generation technology in larger populations are underway.
It bears acknowledgement that the ideal method for deploying AI-ECG analysis in clinical settings continues to be closely studied. Broadly, deployments could focus on either ‘safety net’ or ‘gatekeeper’ applications. In the former, AI-ECG analysis is used to trigger extra echocardiography that may not otherwise be recommended as part of opportunistic screening. Such an approach may improve population-level identification of cardiac disease and may be particularly useful in underserved patient groups at risk for undertesting. In a gatekeeper strategy, the results of an AI-ECG analysis could be used to determine whether patients should undergo echocardiography when clinicians pretest probability is below a certain threshold to try and prevent unnecessary testing. These two strategies have markedly different goals and implications, and the ideal statistics—sensitivity, specificity, positive and negative predictive values—in which to measure their success are sure to differ. Balancing of these statistics may also vary across clinical scenarios. For instance, negative predictive value may be the most important metric in a symptomatic emergency department patient, whereas a deployment focusing on asymptomatic outpatients may focus on maintenance of a high positive predictive value at the cost of a modest sensitivity. As a result of the very high evidence bar needed to overrule a clinician’s judgement to pursue echocardiography given the extensive information available in the history and physical exam, we anticipate that early successful deployments will focus on safety net opportunistic screening rather than forms of gatekeeper functions. More investigation will be required to determine the cost-effectiveness of opportunistic screening approaches using AI models.
On the other hand, there may be potential harms and/or biases associated with using AI models for screening. For example, one potential harm is increased patient anxiety associated with a high-risk prediction, particularly in instances that are ultimately false positives. Arbitrary bias across technicians or physicians is possible, potentially biasing sceptics against diagnosis while biasing enthusiasts toward it. However, over the longer term, this model should evolve to become indistinguishable from other technological advances in medicine and help to ‘normalize’ and thus mitigate these concerns. Nonetheless, further studies on these topics are warranted.
Relationship to previous work
Comparing the accuracy of different deep learning models across distinct healthcare datasets is an enormous challenge, with different patient characteristics having a significant impact on statistical metrics such as AUROC and AUPRC. Other ECG-based SHD models, such as rECHOmmend, have demonstrated excellent performance with an AUROC of 91% in its retrospective dataset4. Owing to the lack of a shared common dataset and differences in SHD definitions (for instance, mild to moderate valve disease being classified as moderate in rECHOmmend and mild in EchoNext) and prevalence (17.9% for rECHOmmend versus 36.3% for EchoNext due to differing exclusion criteria), these results are not directly comparable. For example, the performance of rECHOmmend was observed to drop (AUROC 0.88) when restricting to patients with echocardiography-confirmed disease, thereby increasing disease prevalence, as was done for the present study. Moreover, the SHD label in the current study captures 98.9% of all echo-based diagnoses as compared to 65.6% by rECHOmmend. Compared to other models that have been developed for similar pathologies, the current study had significantly increased racial and/or ethnic diversity across many internal or external institutes. When taken as a whole, this current analysis in conjunction with previously published work demonstrates that deep learning analysis can accurately detect SHDs individually and as a composite, with high accuracy across diverse populations in several clinical contexts and geographically distinct external test sets.
Limitations
Several limitations of our study should be mentioned. Certain labels required binary cutoffs to be arbitrarily decided for what are otherwise continuous values, such as LVEF and low left ventricular wall thickness. Recent registries and trials have defined heart failure with moderately reduced ejection fraction with an upper threshold of 55%, 52.5%, 50% and 45%. The threshold chosen for this study was 45% and results may have differed if a different cutoff was chosen. Supplementary Table 3 includes the performance of the model for only ‘severe’ disease of all the pathologies (for example, LVEF less than or equal to 35% instead of 45%). Restricting the model to a different arbitrary threshold showed a similar AUROC of 87.7% providing evidence that performance was not dependent on any specific threshold for defining disease. The AUPRCs presented in this study are based on retrospective data in which patients have both ECGs and echocardiograms, which is a population with a much higher prevalence of SHD, and thus not representative of AUPRCs we would expect to see if this model was used as a screening study among patients with ECGs and no echocardiograms. Because it is not well known what the underlying prevalence of undiagnosed SHD among patients with ECGs and no echocardiograms, we modelled a range of potential prevalences and the resulting performance of the model in Table 3.
Performance for some component labels was suboptimal. For example, low left ventricular wall thickness is a highly prevalent label but subjective to high interobserver variation, thus introducing inherent label noise. Rare conditions such as pulmonary regurgitation are under-represented and are highly correlated with other more prevalent conditions, such as tricuspid regurgitation max velocity (Supplementary Fig. 4), which makes it hard to learn patterns that are exclusive to pulmonary regurgitation. Furthermore, because we took a multilabel approach to train all labels in one model to best capture collinearity, the model optimization was regularized to minimize overall loss across all labels, performance on certain labels might be compromised to ensure better over performance. The multilabel approach helps to best capture the collinearity and correlation among component disease labels and provides insight on the specific disease(s) that leads to a high-risk prediction result, although not conclusive. Further investigations are needed for specific disease discrimination.
The DISCOVERY trial prospectively recruited patients to assess how the model may perform in general clinical use. The patients were recruited using an earlier generation model, ValveNet, which was only trained to detect left-sided VHD. After the trial completion, EchoNext model performance was assessed on the 100 patients, none of whom were included in model training, validation or testing. Written consent was obtained from all participants. This small trial showed promising results, but larger, more pragmatic interventions will be necessary to determine the potential benefit in excess of usual clinical care for EchoNext.
In conclusion, the EchoNext 12-lead ECG model can accurately detect a composite of SHD across a range of clinical and geographic settings, and the release of the underlying dataset of ECGs with clinically relevant labels can serve as a benchmark for model comparison and further innovation. Future work focusing on deployment strategies is needed to determine whether deep-learning-assisted ECG analysis can be used to improve the diagnosis and outcomes of SHD in clinical practice.
Methods
Patient identification and data sources
Patients 18 years of age or older who underwent a digitally-stored 12-lead ECG between December 2008 and 2022 at one of eight NYP-affiliated hospitals (Columbia University Irving Medical Center, Weill Cornell Medical Center, NYP-Brooklyn Methodist Hospital, NYP-Lower Manhattan Hospital, NYP-Queens Hospital, NYP-Allen Hospital, NYP-Westchester Hospital and adult patients at the Morgan Stanley Children’s Hospital of New York) were identified. Those 230,318 unique patients who had a diagnostic quality, non-ventricularly paced ECG performed up to 1 year before an echocardiogram formed the NYP multicentre cohort, with 1,245,273 distinct ECG–echocardiogram pairs (Fig. 1). Demographics, including race and ethnicity, were abstracted from the electronic medical record.
ECG data were accessed from the MUSE data management system (GE Healthcare) at each institution. ECG data that were abstracted included demographic and ECG-specific tabular information, including age, sex, atrial and ventricular rates, and pulmonary regurgitation, QRS and Bazett’s corrected Q wave-to-T wave intervals. The ECG waveform data were abstracted at 250 Hz for all 12 ECG leads for a total of 30,000 data points.
Echocardiographic data were accessed from the Syngo Dynamics (Siemens Healthineers) and Xcelera (Phillips) systems. Abstracted data included the LVEF, interventricular septum and posterior wall thicknesses (with the larger being defined as the maximum low left ventricular wall thickness), qualitative right ventricular systolic function (defined as normal, mildly reduced, moderately reduced or severely reduced), the PASP and maximum tricuspid regurgitation maximum velocity, the presence of a pericardial effusion (normalized to a scale of none or trace, small, moderate or large) and the severity of the VHDs of aortic stenosis, aortic regurgitation, mitral regurgitation, tricuspid regurgitation and pulmonic regurgitation (normalized to a scale of none or trace, mild, moderate or severe). Mild to moderate VHD was classified as mild disease and moderate to severe VHD was classified as moderate disease. Repaired or replaced heart valves were excluded from the dataset. These data were harmonized across each echocardiographic reading system and hospital with a minimum of 100 cases audited per label to confirm the accuracy of each analysis. From these features, we defined the presence or absence of SHD for each echocardiogram using the following binary cutoffs: LVEF (less than or equal to 45%), maximum low left ventricular wall thickness (greater than or equal to 1.3 cm), right ventricular dysfunction (moderate or severely reduced), pulmonary hypertension (PASP greater than or equal to 45 mm Hg or tricuspid regurgitation jet velocity greater than or equal to 3.2 m s−1), aortic stenosis (moderate or severe), aortic regurgitation (moderate or severe), mitral regurgitation (moderate or severe), tricuspid regurgitation (moderate or severe), pulmonary regurgitation (moderate or severe) and a significant pericardial effusion (moderate or large). For an ECG to be labelled as being ‘positive’ for a disease, it must have been performed within 1 year before an echocardiogram with SHD. In patients without SHD (confirmed by at least one ‘negative’ echocardiogram), all ECGs before the most recent echo were labelled as negative and included in the study. Only ECGs with an echo occurring afterwards were used to ensure no ECGs occurring after corrective procedures in which a future echo may not occur were included as they would be mislabelled.
To be included in the study, an echocardiography report was required to include LVEF, a wall thickness measurement and one relevant valve finding. Missing data were imputed using the following process. For valve findings, if either regurgitation or stenosis was commented on, the other was presumed normal (for example, if aortic stenosis was reported but aortic regurgitation was not, then we assumed no aortic regurgitation). If not specifically commented on, a pericardial effusion and pulmonary hypertension were presumed to be absent.
In addition to this base SHD label, a secondary, more stringent cutoff was defined for each endpoint (for example, LVEF less than or equal to 35%) to reflect ‘severe SHD’. These cutoffs and model accuracy using these severe SHD endpoints are detailed in the Supplementary Information.
For the primary analysis, data from all eight hospital campuses were blended and split by patient into training, validation and test sets (64%, 16% and 20%). Further experiments are detailed in the Supplementary Information; for example, using alternate data partitions to hold out specific NYP hospitals from training to assess generalization. In all cases, several ECG–echocardiogram pairs were used in training, but the most recent ECG–echocardiogram pair was selected for each unique patient in the validation and test sets. This retrospective study was conducted with approval of the Columbia University and Weill Cornell Institutional Research Boards with waiver of patient consent.
Model details
The EchoNext model comprises a convolutional neural network that takes a digital 12-lead ECG waveform, patient demographics and ECG-specific tabular information to predict the presence or absence of SHD (Supplementary Table 1). Extending previous work3, we trained EchoNext as a multitask classifier such that separate terminal branches of the model predict the presence of the SHD composite label and the presence of an individual component label (for instance, the presence or absence of aortic stenosis), respectively. Details on the model design, hyperparameters, testing and optimization are addressed in the Supplementary Information.
Silent deployment validation
As the model development dataset comprised ECGs acquired throughout December 2022, we subsequently collected ECG–echocardiogram pairs acquired at NYP from January to 16 September 2023, as a temporally distinct validation set. Patients included in the development cohort were excluded from this analysis.
Prospective validation of ValveNet and EchoNext
Before development of EchoNext, study investigators had created ValveNet, a similarly architected AI-ECG model trained to detect the left-sided VHD of aortic stenosis, aortic regurgitation and mitral regurgitation, a subset of SDH3. To test the ability of a system using this model to detect clinically significant cardiac disease, we designed the Aortic Stenosis Discovery Study, a 100-patient, open-label trial. Adult patients were eligible if they had a digital 12-lead ECG performed at Columbia University and had no history of an echocardiogram within the last 3 years in our system, no history of left-sided VHD and no dementia or other non-cardiac life-limiting disease with expected survival less than 1 year. Eligible patients were recruited by their ValveNet score (a continuous variable from 0–1 with a value closer to 1 indicating a higher model confidence that VHD was present) into high-risk (score greater than or equal to 0.6) or moderate-risk (score 0.3–0.6) groups. Patients with scores less than 0.3 were excluded due to a very low predicted risk of cardiac disease. Consented patients underwent an echocardiogram. The primary endpoint was moderate or severe aortic stenosis, aortic regurgitation or mitral regurgitation. The key secondary endpoint was any SHD that was identical to the EchoNext label. Critically important findings were communicated with patients and physicians, and appropriate clinical follow-up was coordinated by study investigators for newly diagnosed disease.
Cardiologist survey
Board-certified attending cardiologists were recruited from Columbia University to study human accuracy in the detection of SHD using the ECG. A total of 13 cardiologists were recruited to take this study (J.M.D., S.Y., G.F.R., S.R.A., Q.L., C.K.B., P.V., C.A.W., E.M.D., V.A., M. Lebehn, P.N.K. and S.S.). A total of 150 ECGs were selected from the NYP multicentre test set representing a similar age distribution and SHD prevalence to the entire dataset. The digital ECG was accessed as a pdf and the name, date and clinical interpretation was cropped out of the image leaving only the waveform and the ECG measurements (ventricular rate, pulmonary regurgitation interval, QRS interval, Q wave-to-T wave interval and axis). The age (truncated to greater than 90) and sex were added to each ECG. These 150 ECGs were split into blocks of 50. Each cardiologist was presented with a block of 50 ECGs and were asked to answer two questions for each ECG: whether the patient was likely to have SHD (not likely or likely). After completion of each block of 50 ECGs, they were given the same 50 ECGs with the addition of the AI model analysis with both the model output (0–1) and model interpretation (less than 0.6 not consistent with SHD, greater than or equal to 0.6 consistent with SHD) added to the image. Each cardiologist could complete up to 300 ECGs (150 without and 150 with the AI model analyses). The results from all the cardiologists were pooled for primary analysis with calculation of the accuracy, sensitivity and specificity using standard methods with 95% CIs for accuracy calculated by the Clopper–Pearson method. The accuracy of the EchoNext model in this 150 ECG dataset was determined using a threshold of 0.6. This method of human–machine comparison is similar to typical methods and one we have used previously34,35. Clinically normal ECGs were identified from the clinical interpretation report and performance was compared between normal and abnormal ECGs.
Statistical analysis
Descriptive statistics were used to describe the data using standard methods. The performance of EchoNext was assessed using standard metrics, including AUROC and AUPRC. Diagnostic odds ratio was also computed at the operating point of 0.5. For each statistical test, 95% CIs were generated using 1,000 bootstrapped estimates. Subgroup analyses were performed using subsets of age, sex, race and ethnicity. All statistical analyses were performed using Python v.3.8.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
To facilitate future research and innovation in this field, a de-identified 100,000-patient subset of the NYP multicentre cohort (specific to Columbia University Medical Center) will be released with publication of this study. This dataset will contain all input features needed to run the model (ECG waveforms, tabular ECG features and patient demographics), as well as the corresponding disease labels derived from echocardiography. To provide a benchmark for future comparison, we split this dataset into train, validation and test, and report the performance of the model within the public dataset. These data are available at GitHub (https://github.com/PierreElias/IntroECG).
Code availability
The Columbia mini-model was developed using the training and validation data splits provided in the publicly released Columbia ECG dataset (Supplementary Table 13). Reported performance is based on the public test set (Supplementary Table 15); performance was also assessed in the NYP multicentre cohort for direct comparison with the EchoNext model (Supplementary Table 14). As with EchoNext, this is a multitask model that predicts both the composite SHD label and a component disease. Performance for each individual disease component is included in Supplementary Tables 14 and 15. The code used to train and evaluate the model, and the specific model weights are also publicly available for non-commercial use, as detailed above. The code is available at GitHub (https://github.com/PierreElias/IntroECG).
References
Otto, C. M. et al. 2020 ACC/AHA guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 143, e72–e227 (2021).
Heidenreich, P. A. et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 79, 1757–1780 (2022).
Elias, P. et al. Deep learning electrocardiographic analysis for detection of left-sided valvular heart disease. J. Am. Coll. Cardiol. 80, 613–626 (2022).
Ulloa-Cerna, A. E. et al. rECHOmmend: an ECG-based machine learning approach for identifying patients at increased risk of undiagnosed structural heart disease detectable by echocardiography. Circulation 146, 36–47 (2022).
Siontis, K. C., Noseworthy, P. A., Attia, Z. I. & Friedman, P. A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 18, 465–478 (2021).
2020 Heart Disease and Stroke Statistical Update Fact Sheet At-a-Glance (American Heart Association, 2020); www.heart.org/-/media/files/about-us/statistics/2020-heart-disease-and-stroke-ucm_505473.pdf?la=en#:~:text=In%202017%2C%20Coronary%20Heart%20Disease,other%20cardiovascular%20diseases%20(17.6%25).
Tsao, C. W. et al. Heart disease and stroke statistics-2023 update: a report from the American Heart Association. Circulation 147, e93–e621 (2023).
Mallow, P. J., Chen, J., Moore, M., Gunnarsson, C. & Rizzo, J. A. Incremental direct healthcare expenditures of valvular heart disease in the USA. J. Comp. Eff. Res. 8, 879–887 (2019).
Urbich, M. et al. A systematic review of medical costs associated with heart failure in the USA (2014–2020). Pharmacoeconomics 38, 1219–1236 (2020).
Disease, G. B. D., Injury, I. & Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1789–1858 (2018).
Mensah, G. A. et al. Global burden of cardiovascular diseases and risks 1990-2022. J. Am. Coll. Cardiol. 82, 2350–2473 (2023).
Stierman, B. et al. National Health and Nutrition Examination Survey 2017–March 2020 Prepandemic Data Files—Development of Files and Prevalence Estimates for Selected Health Outcomes. Report NHSR No. 158 (Centers for Disease Control and Prevention, National Center for Health Statistics, 2021).
Osnabrugge, R. L. et al. Aortic stenosis in the elderly: disease prevalence and number of candidates for transcatheter aortic valve replacement: a meta-analysis and modeling study. J. Am. Coll. Cardiol. 62, 1002–1012 (2013).
d’Arcy, J. L. et al. Large-scale community echocardiographic screening reveals a major burden of undiagnosed valvular heart disease in older people: the OxVALVE Population Cohort Study. Eur. Heart J. 37, 3515–3522 (2016).
Kang, D. H. et al. Early surgery or conservative care for asymptomatic aortic stenosis. N. Engl. J. Med. 382, 111–119 (2020).
Suri, R. M. et al. Association between early surgical intervention vs watchful waiting and outcomes for mitral regurgitation due to flail mitral valve leaflets. JAMA 310, 609–616 (2013).
Cohen-Shelly, M. et al. Electrocardiogram screening for aortic valve stenosis using artificial intelligence. Eur. Heart J. 42, 2885–2896 (2021).
Kwon, J. M. et al. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J. Am. Heart Assoc. 9, e014717 (2020).
Adedinsewo, D. et al. Artificial intelligence-enabled ECG algorithm to identify patients with left ventricular systolic dysfunction presenting to the emergency department with dyspnea. Circ. Arrhythm. Electrophysiol. 13, e008437 (2020).
Ko, W. Y. et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J. Am. Coll. Cardiol. 75, 722–733 (2020).
Tison, G. H. et al. Assessment of disease status and treatment response with artificial intelligence−enhanced electrocardiography in obstructive hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 79, 1032–1034 (2022).
Baumgartner, H. et al. Recommendations on the echocardiographic assessment of aortic valve stenosis: a focused update from the European Association of Cardiovascular Imaging and the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 30, 372–392 (2017).
Lang, R. M. et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 28, 1–39 e14 (2015).
Mitchell, C. et al. Guidelines for performing a comprehensive transthoracic echocardiographic examination in adults: recommendations from the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 32, 1–64 (2019).
Zoghbi, W. A. et al. Recommendations for noninvasive evaluation of native valvular regurgitation: a report from the American Society of Echocardiography developed in collaboration with the Society for Cardiovascular Magnetic Resonance. J. Am. Soc. Echocardiogr. 30, 303–371 (2017).
Guazzi, M. & Borlaug, B. A. Pulmonary hypertension due to left heart disease. Circulation 126, 975–990 (2012).
Merlos, P. et al. Echocardiographic estimation of pulmonary arterial systolic pressure in acute heart failure. Prognostic implications. Eur. J. Intern. Med. 24, 562–567 (2013).
Solomon, S. D. et al. Angiotensin–neprilysin inhibition in heart failure with preserved ejection fraction. N. Engl. J. Med. 381, 1609–1620 (2019).
Armstrong, P. W. et al. Vericiguat in patients with heart failure and reduced ejection fraction. N. Engl. J. Med. 382, 1883–1893 (2020).
Savarese, G., Stolfo, D., Sinagra, G. & Lund, L. H. Heart failure with mid-range or mildly reduced ejection fraction. Nat. Rev. Cardiol. 19, 100–116 (2022).
Attia, I. Z. et al. External validation of a deep learning electrocardiogram algorithm to detect ventricular dysfunction. Int. J. Cardiol. 329, 130–135 (2021).
Fisch, C. Centennial of the string galvanometer and the electrocardiogram. J. Am. Coll. Cardiol. 36, 1737–1745 (2000).
Fye, W. B. A history of the origin, evolution, and impact of electrocardiography. Am. J. Cardiol. 73, 937–949 (1994).
Oakden-Rayner, L. & Palmer, L. J. in Artificial Intelligence in Medical Imaging: Opportunities, Applications and Risks (eds Ranschaert, E. R. et al.) 83–104 (Springer, 2019).
Bhave, S. et al. Deep learning to detect left ventricular structural abnormalities in chest X-rays. Eur. Heart J. 45, 2002–2012 (2024).
Acknowledgements
T.J.P. has research support provided to his institution from the Amyloidosis Foundation and the American Heart Association (grant award nos. 933452 and 23SCISA1077494; https://doi.org/10.58275/AHA.23SCISA1077494.pc.gr.172160). M.A.P. is at present supported by an R01 grant from the National Heart, Lung, and Blood Institute of the US National Institutes of Health (grant no. R01HL149680). B.D. is at present supported by financial support from the Patient Centered Outcomes Research Institute (grant no. PCORI IHS-2019C2-17373-IC). R.A. is supported by the ‘Fonds de la recherche en santé du Québec’ (grant no. 312758), the Montreal Heart Institute Research Centre, the Montreal Heart Institute Foundation and the Des Groseillers-Bérard Research Chair. P.E. has research support provided to his institution from the American Heart Association and the New York Academy of Medicine.
Ethics declarations
Competing interests
Columbia University has submitted a patent application (no. 63/555,968) on the EchoNext ECG algorithm, on which T.J.P., L.J., C.M.H. and P.E. are inventors. T.J.P. owns stock in Abbott Laboratories and Baxter International, with research support provided to his institution from Eidos Therapeutics, Janssen, Pfizer and Edwards Lifesciences. E.M.D. serves on a clinical trial committee for Abiomed. R.T.H. has institutional consulting contracts with Abbott Structural, Edwards Lifesciences, Medtronic and Novartis, for which she receives no direct compensation. M.A.P. received a one-time research donation from Roche Diagnostics in 2023. B.D. is a consultant for Hone Health. R.A. and G.H.T. are co-inventors on the patent 63/208,406 (Method and System for Automated Analysis of Coronary Angiograms). R.A. reports receiving speaker fees from Abbott, Boston Scientific, Boehringer-Ingelheim and Novartis. G.H.T. is an adviser to Prolaio and Viz.ai, and has received research grants from Janssen Pharmaceuticals, General Electric and Myokardia, a wholly owned subsidiary of Bristol Meyers Squibb. P.E. has research support provided to his institution from Eidos Therapeutics, Pfizer, Janssen, Edwards Lifesciences and Google. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Mintu Turakhia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Poterucha, T.J., Jing, L., Ricart, R.P. et al. Detecting structural heart disease from electrocardiograms using AI. Nature (2025). https://doi.org/10.1038/s41586-025-09227-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41586-025-09227-0