- Article
- Open access
- Published:
Scientific Reports volume 15, Article number: 29248 (2025) Cite this article
Abstract
The solubility of medications in supercritical solvent is the most important factor that can be determined via appropriate computational tools. This work explores the modeling of digitoxin solubility as the case study in supercritical CO2 and solvent density utilizing ensemble methods. Temperature and pressure are the input parameters, while solvent density and digitoxin solubility are the output parameters. Several machine learning models along with optimizer were used for correlation of the dataset. Employing AdaBoost as an ensemble method, predictions from Bayesian Ridge Regression (BRR), Gaussian process regression (GPR), and K-nearest neighbors (KNN) are amalgamated. Sailfish Optimizer (SFO) is utilized for hyper-parameter tuning to enhance model performance. Results reveal that AdaBoost combined with ADA-GPR exhibits the lowest Average Absolute Relative Deviation (AARD%) values, with solubility achieving 7.74 and solvent density reaching 2.76, respectively. This underscores the efficacy of ensemble methods and hyper-parameter tuning in accurately predicting complex chemical properties in supercritical CO2 systems.
Introduction
To enhance the solubility of medicines, size reduction is the most commonly used method which can be implemented for various solid-dosage formulations. Those drugs that are classified as BCS class II, possess low solubility in aqueous solutions. The drugs with low solubility and high permeability are classified in this group according to Biopharmaceutical Classification System (BCS)1. Size reduction will enhance the solubility and consequently the bioavailability owing to the higher surface energy of drug powder in the nanosize2. Therefore, this approach is suitable in the pharmaceutical industry for improving the properties of medications with poor solubility. Another approach for enhancing the solubility is amorphization by which a drug is transformed from crystalline to amorphous state with higher solubility. However, the problem with the amorphous state is its poor stability which recrystallizes over time and loses the properties of amorphous state. As such, it must be stabilized which can be done via distribution of drugs inside a polymeric matrix to prevent mobility and recrystallization3,4.
For size reduction, supercritical method can be employed, which is a newly developed process for pharmaceutical processing where it has the ability to reduce the size of particles in a continuous model of operation. The method is also mentioned as green processing due to the lack of organic solvent in the size reduction operation, thereby offering sustainable processing5,6,7. For size reduction using this method, the drug must be dissolved in the solvent at an acceptable value, thus the solubility of drug in supercritical solvent is the limiting step for development of this green process. As such, the solubility of medicine in the supercritical solvent should be estimated via robust methods to ensure the reliability of method in precise determination of solubility prior to the operation. Computational techniques can be used as screening tools for assessment of drugs nanoparticle production in this process. Thermodynamic approach is a reliable tool for estimating drug solubility in supercritical solvents (e.g., CO2), which is built based on phase equilibria, such as solid-liquid equilibrium8,9.
Despite the advantages of thermodynamic models for correlating medicines solubility in supercritical solvents, the models are not easy to be applied and there is sometimes complexity in implementing these models. As such, development of models with less complexity and generality would be preferred for estimating drugs solubility in supercritical solvents. Data-driven approach is an alternative for correlation of drugs solubility which rely on the availability of measurements and dataset. Some machine learning and optimizer algorithms have been implemented for estimation of drugs solubility in supercritical carbon dioxide, and reports have shown the models are of great accuracy in this field10,11,12,13. One of the data-driven models is machine learning (ML) where its domain is currently undergoing significant growth and has been widely employed in various applications, including the analysis of measured data for the purpose of optimization, regression, and prediction14. AI (Artificial Intelligence) based models and deep learning (DL) have been recently studied for pharmaceutical and energy applications15,16,17 which can be further explored for solubility analysis of medications.
Machine learning techniques facilitate the creation of accurate and reliable models that can identify patterns within complex datasets and produce predictions based on those patterns18,19,20. This study utilizes AdaBoost as an ensemble method to incorporate predictions from Gaussian process regression (GPR), Bayesian Ridge Regression (BRR), and K-nearest neighbors (KNN). The Sailfish Optimizer (SFO) is employed for the purpose of hyper-parameter tuning in to enhance the performance of the model.
In the field of Bayesian theory and statistical learning theory, GPR has emerged as a useful machine learning technique. The framework provides a flexible structure for executing probabilistic regression and is widely utilized for tackling regression issues marked by high dimensionality, limited sample sizes, or nonlinearity21. Bayesian Ridge Regression models are a combination of Bayesian inference and regression techniques. This method provides more reliable and efficient estimation of coefficients in linear models compared to least squares regression22. The KNN regression model is a data-driven technique employed for the purpose of predicting short-term traffic flow. The algorithm operates by identifying the K most analogous instances to a provided input and calculating their mean as the prediction23.
So, the current study develops a methodology based on ML for modeling drug solubility in supercritical CO2 and makes correlation with pressure and temperature. The models included GPR, BRR, and KNN which are optimized using Sailfish Optimizer. Digitoxin is selected in this study for implementing methodology and assessing the reliability of models and optimizer.
Materials and methods
Data of digitoxin solubility
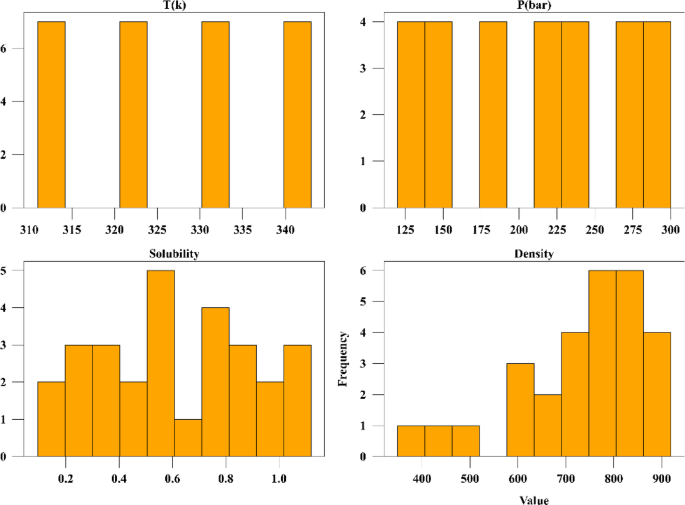
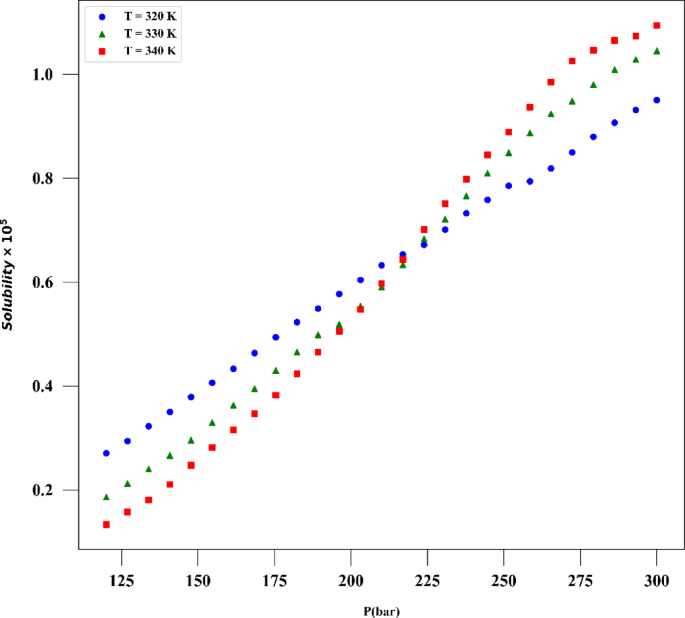
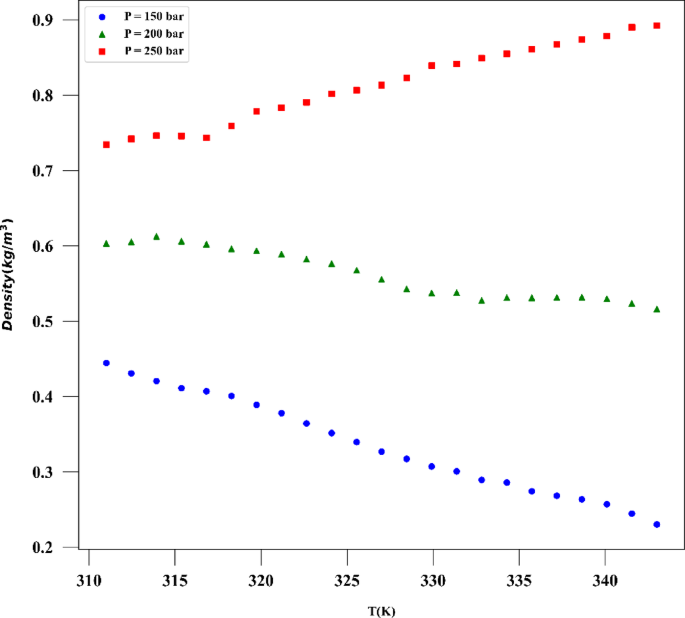
A number of measured data was applied for developing and testing the models which are collected from reference24 and its statistical analysis is presented in Table 1 which are based on raw data. The drug used for the analysis is digitoxin, and its solubility in supercritical CO2 was collected and used in this work for building ML models and optimizer. The same data was already used by Li et al.25 to build several ML techniques in estimation of digitoxin solubility. Temperature and pressure are the two input features in this dataset owing to their significant impact on the solubility of digitoxin. As the solvent is compressible, the influence of pressure should be evaluated to understand its effect on digitoxin solubility changes. Figure 1 shows histograms of all parameters of the dataset. Thus, the machine learning models were developed utilizing two inputs and two outputs which are solubility of drug and the density of solvent.
Sailfish optimizer (SFO)
Enhancing the effectiveness of machine learning algorithms hinges crucially on optimizing hyper-parameters. The choice of appropriate hyper-parameters profoundly influences both the accuracy and the generalization capabilities of these algorithms26. In the last several years, there has been escalating interest in nature-inspired optimization methods, owing to their efficacy in fine-tuning hyper-parameters27. The Sailfish Optimizer (SFO) is an algorithm that is influenced by the synchronized swimming behavior observed in sailfish28.
The SFO algorithm is a metaheuristic method inspired by the hunting and cooperative behavior of marine sailfish29. Mathematically, the Sailfish Optimizer functions by iteratively adjusting the hyper-parameters through a series of equations that mimic the motion of the sailfish. The equation governing the update of the position of the i-th sailfish in D-dimensional space can be expressed as follows28:
$$\:{X}_{i}^{t+1}={X}_{i}^{t}+{V}_{i}^{t+1}$$
where \(\:{X}_{i}^{t}\) stands for the position of the i-th sailfish at t-th iteration, and\(\:{V}_{i}^{t+1}\) denotes the velocity vector of the i-th sailfish at the next iteration28.
ADABOOST
AdaBoost, an abbreviation for “adaptive boosting,” is a widely utilized machine learning approach that surpasses simpler algorithms in predictive accuracy. This method involves adjusting a weighted combination of functions to align with the aggregated data, determining the total error, and then modifying an initial function accordingly. At each step, multiple base models are implemented on the adjusted dataset, and their respective errors are assessed. Consequently, the mistake from one iteration influences the subsequent model in every cycle. The final prediction is derived by assigning appropriate weights to each function and summing their contributions once the adjusted error is minimized to zero30,31.
Base models
We utilized Gaussian Process Regression (GPR) as one of the base models. A Gaussian process (GP) is a collection of random variables some of which have Gaussian distributions (GDs)32,33. The covariance and mean functions serve as effective metrics for evaluating the performance of a GP. In the GPR framework, GDs are extended, with the mean represented as a vector and the covariance as a matrix34,35.
What distinguishes GPR from other regression models is its lack of necessity for a precise specification of a fitting function. Instead, it utilizes statistical models that approximate a random sample from a multidimensional GD to interpret field data34.
The second base model is Bayesian Ridge Regression (BRR). BRR combines Bayesian inference with regression models. This method estimates coefficients in linear models more robustly and efficiently than least squares regression. BRR estimates linear model coefficients using prior data beliefs. A prior distribution encodes prior beliefs, which are integrated with the likelihood function to create the posterior distribution. Estimating linear model coefficients using this distribution allows data estimation. We assume a normal distribution of regression coefficients with a zero mean and an alpha hyper-parameter that determines accuracy (inverse variance).
The likelihood function follows a normal distribution, where the linear regression model predicts the mean and the additional hyperparameter, lambda, determines the variance. The aim is to ascertain the probable values of the regression coefficients β by leveraging existing data and prior information. The subsequent equation defines the posterior distribution of β36:
$$\:p(\beta\:\mid\:X,y,\alpha\:,\lambda\:)=\text{N}(\beta\:\mid\:\mu\:,\varSigma\:)$$
The mean vector represented by \(\:\mu\:\) and the covariance matrix \(\:\varSigma\:\) in the provided equation denote the statistical properties of the posterior distribution. Analytical calculations are performed using the Bayesian formula to determine the values of these parameters37:
$$\:\mu\:={(\lambda\:\cdot\:{X{\prime\:}X}^{\cdot\:}+\alpha\:\cdot\:I)}^{-1}\cdot\:{X{\prime\:}y}^{\cdot\:}$$
$$\:\varSigma\:={(\lambda\:\cdot\:{X{\prime\:}X}^{\cdot\:}+\alpha\:\cdot\:I)}^{-1}$$
In the context mentioned, \(\:X{\prime\:}X\) stands for the result of transposing the matrix of independent variables and then multiplying it by itself, while \(\:X{\prime\:}y\) denotes the product of transposing the matrix of independent variables (input) and multiplying it by the matrix of dependent variables (output). Additionally, the symbol \(\:I\) denotes the identity matrix. More details are reported elsewhere about the model38,39,40.
Results and discussion
The models implementation was performed using Python software, 3.8 version, accessible at: https://www.python.org. The hyperparameters of the proposed models were optimized via SFO algorithm as previously outlined. The optimized models were used for evaluation of drug solubility and finding which model can best predict digitoxin solubility in the supercritical solvent. The results obtained for each regression model in terms of Average Absolute Relative Deviation (AARD%), Root Mean Square Error (RMSE), and Mean Absolute Error (MAE) are summarized in Tables 2 and 3 for both solubility and density, respectively. The errors between the estimated and measured values have been calculated and used as the main metrics for evaluation of the models’ precision.
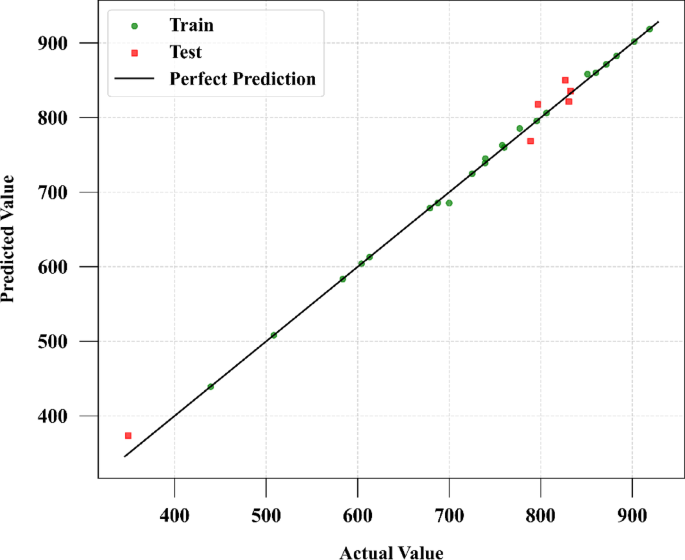
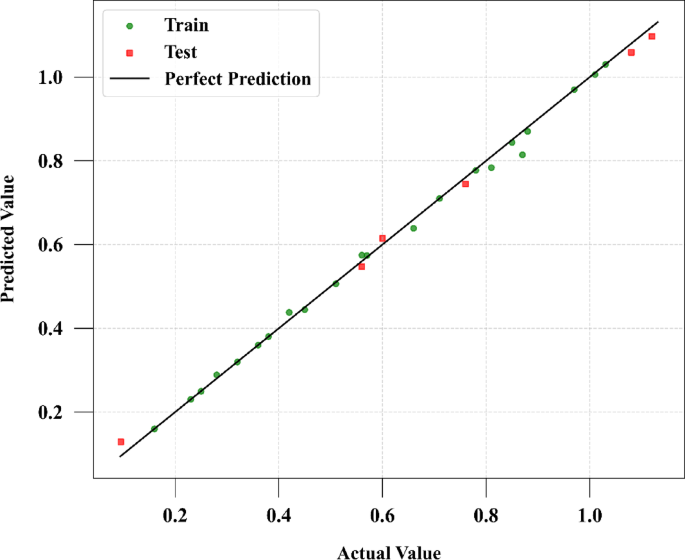
The findings reveal that the ADA-GPR model outperforms the other models in terms of solubility and solvent density prediction. Figures 2 and 3 show a comparison of expected and predicted values for both outputs using this model. The ADA-GPR model achieved the lowest AARD% values for both predictions. Specifically, for solubility prediction, ADA-GPR achieved an AARD% of 7.74721E + 00, while for solvent density prediction, it attained an AARD% of 2.76323E + 00. Additionally, the ADA-GPR model demonstrated the lowest RMSE and MAE values, further highlighting its robust performance for estimation of digitoxin solubility.
The comparative analysis of the best model in this work is indicated in Table 4. ADA-GPR model optimized in this study has AARD% of 7.74 which is lower than the traditional thermodynamic models based on Equation of State (EoS) developed by Sheikhi-Kouhsar et al.24.
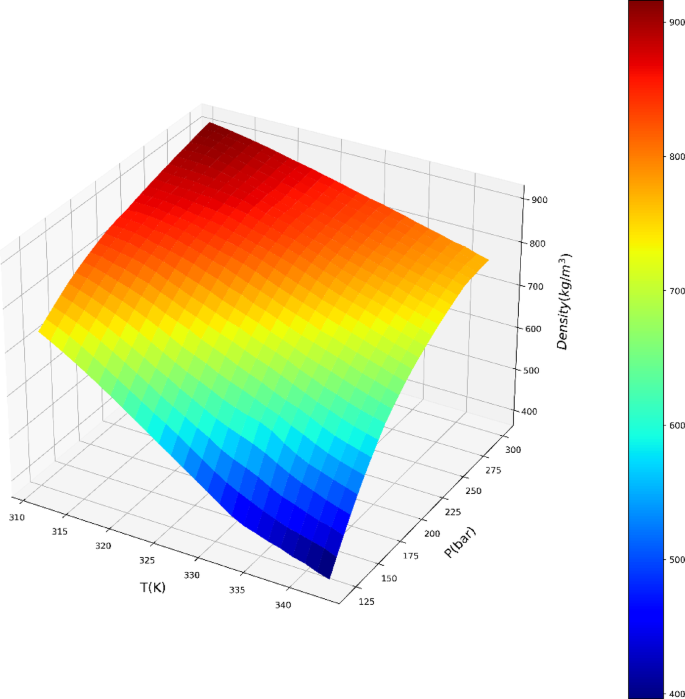
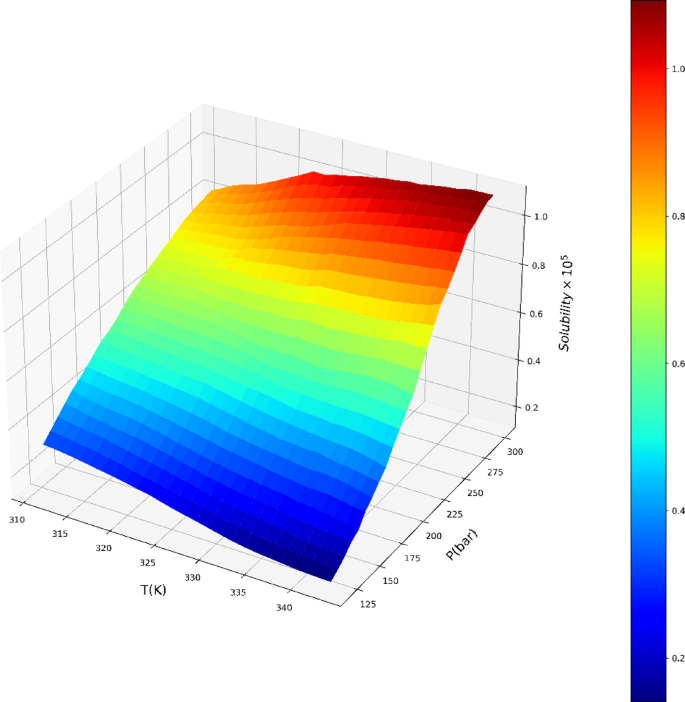
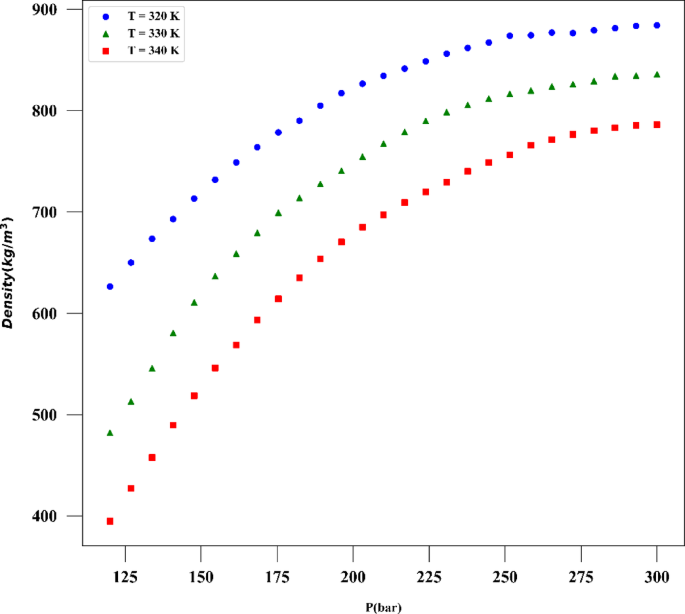
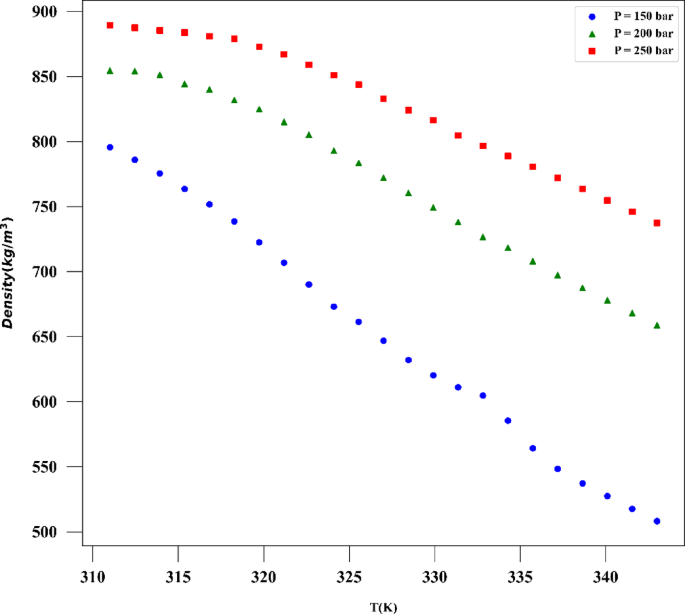
The exceptional efficacy demonstrated by the ADA-GPR model can be ascribed to its adeptness in capturing intricate correlations within the dataset and its adaptability in depicting nonlinear patterns. The ADA-BRR model also performed reasonably well, particularly for solubility prediction, but it is behind the ADA-GPR model in terms of fitting accuracy. On the other hand, the ADA-KNN model exhibited the highest AARD% values and the poorest performance among the three models. The final analysis was conducted using the ADA-GPR model based on the available data. Final prediction surfaces are shown in Figs. 4 and 5. Also, Figs. 6, 7, 8 and 9 are the partial dependencies between inputs and outputs. Confirmation is observed for the results obtained in this research through comparing with the previous studies on computation of pharmaceutical solubility, while the variations have been seen to be similar25. The pressure and temperature effects on the solubility and density in this work show agreement with previous works which reported the use of machine learning in drugs solubility correlation25,41,42,43.
The results confirm the validity of the methodology designed in this work, as the observations match the experimental trends. For the drug solubility versus temperature, a decreasing trend is seen in Fig. 9 for low pressure, while for higher pressure, the solubility increases with temperature. This observation in the drug solubility is related to the cross-over pressure zone which can change the path of solubility variations by enhancing the temperature. As for density, the trend is justified, and one can observe that the density is increased with rising pressure and decreased with increasing T. The trend of pressure is owing to the compressibility of the solvent that is supercritical CO225,44. Thus, it is clearly revealed by the models in this study that the digitoxin solubility in the solvent can be significantly varied by adjusting the pressure as well as temperature to achieve the target value for solubility42. A function can be then defined to reversely estimate the values of pressure and temperature for a given digitoxin solubility by the aid of optimized ML model. Furthermore, the combined effects of density variations on the solubility can be analyzed as the models are able to determine density values versus temperature and pressure.
Conclusion
In this research work, we investigated the prediction of solubility of digitoxin and the density of the solvent in supercritical CO2 utilizing ensemble methods and regression models. We employed the AdaBoost ensemble method to combine the predictions from GPR, BRR, and KNN regression models. Hyper-parameter optimization was conducted utilizing the Sailfish Optimizer (SFO) to optimize the performance of the models. Our findings reveal that the AdaBoost ensemble method combined with GPR yielded the most accurate predictions for both solvent density and solubility, as evidenced by the lowest Average Absolute Relative Deviation (AARD%) values. Specifically, for solubility prediction, the AdaBoost-GPR combination achieved an AARD% of 7.74, while for solvent density prediction, it attained an AARD% of 2.76. These results underscore the effectiveness of ensemble methods and hyper-parameter tuning methods in accurately predicting complex chemical properties in supercritical CO2 systems. The findings obtained from this study have the potential to contribute valuable knowledge to future research efforts focused on enhancing hyper-parameters and refining predictive modeling methods for analogous chemical systems.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Csicsák, D. et al. The effect of the particle size reduction on the biorelevant solubility and dissolution of poorly soluble drugs with different acid-base character. Pharmaceutics 15(1), 278 (2023).
Kumar, R. et al. Particle size reduction techniques of pharmaceutical compounds for the enhancement of their dissolution rate and bioavailability. J. Pharm. Innov. 17(2), 333–352 (2022).
Rams-Baron, M. et al. Amorphous Drug Solubility and Absorption Enhancement, in Amorphous Drugs: Benefits and Challenges 41–68 (Springer International Publishing, 2018).
Zhuo, X. et al. Mechanisms of drug solubility enhancement induced by β-lactoglobulin-based amorphous solid dispersions. Mol. Pharm. 20(10), 5206–5213 (2023).
Abourehab, M. A. S. et al. Theoretical investigations on the manufacture of drug nanoparticles using green supercritical processing: Estimation and prediction of drug solubility in the solvent using advanced methods. J. Mol. Liq. 120559 (2022).
Faris Alotaibi, H. et al. Pharmaceutical nanonization by green supercritical processing: investigation of exemestane anti-estrogenic medicine solubility using machine learning. J. Mol. Liq. 392, 123353 (2023).
Zhang, Y. Analysis of nanonization and purification of organic compounds via green supercritical processing: model development using advanced hybrid techniques. Case Stud. Therm. Eng. 55, 104159 (2024).
Faraz, O. et al. Thermodynamic modeling of pharmaceuticals solubility in pure, mixed and supercritical solvents. J. Mol. Liq. 353, 118809 (2022).
Zarei, A., Haghbakhsh, R. & Raeissi, S. Overview and thermodynamic modelling of deep eutectic solvents as co-solvents to enhance drug solubilities in water. Eur. J. Pharm. Biopharm. 193, 1–15 (2023).
Alanazi, M. et al. Development of a novel machine learning approach to optimize important parameters for improving the solubility of an anti-cancer drug within green chemistry solvent. Case Stud. Therm. Eng. 49, 103273 (2023).
Cenci, F. et al. Predicting drug solubility in organic solvents mixtures: A machine-learning approach supported by high-throughput experimentation. Int. J. Pharm. 660, 124233 (2024).
Ghazwani, M. et al. Development of advanced model for understanding the behavior of drug solubility in green solvents: machine learning modeling for small-molecule API solubility prediction. J. Mol. Liq. 386, 122446 (2023).
Wang, C. et al. Prediction of enhanced drug solubility related to clathrate compositions and operating conditions: machine learning study. Int. J. Pharm. 646, 123458 (2023).
Rabbani, Y. et al. Application of artificial neural networks and support vector regression modeling in prediction of magnetorheological fluid rheometery. Colloids Surf., A. 520, 268–278 (2017).
Togun, H. et al. Advancing organic photovoltaic cells for a sustainable future: the role of artificial intelligence (AI) and deep learning (DL) in enhancing performance and innovation. Sol. Energy. 291, 113378 (2025).
Halawani, R. F. et al. An advanced heat design-CO2 capture network for an oxy-biogas fuel combustion cycle combined with a CAES-based method for peak shaving: An artificial intelligent-driven optimization. Renew. Energy. 242, 122474 (2025).
Saini, J. P. S., Thakur, A. & Yadav, D. AI-driven innovations in pharmaceuticals: optimizing drug discovery and industry operations. RSC Pharm. 2(3), 437–454 (2025).
Alpaydin, E. Introduction To Machine Learning (MIT Press, 2020).
Graish, M. S. et al. Prediction of the viscosity of iron-CuO/water-ethylene glycol non-Newtonian hybrid nanofluids using different machine learning algorithms. Case Stud. Chem. Environ. Eng. 11, 101180 (2025).
Hajinajaf, N. et al. Integrated CO2 capture and nutrient removal by microalgae chlorella vulgaris and optimization using neural network and support vector regression. Waste Biomass Valoriz. 13(12), 4749–4770 (2022).
Zhikun, H. et al. Overview of Gaussian process regression. Control Decis. 28(8), 1121–1129 (2013).
Shi, Q., Abdel-Aty, M. & Lee, J. A Bayesian ridge regression analysis of congestion’s impact on urban expressway safety. Accid. Anal. Prev. 88, 124–137 (2016).
Kang, S. K-nearest neighbor learning with graph neural networks. Mathematics 9(8), 830 (2021).
Sheikhi-Kouhsar, M. et al. Solubility of digitoxin in supercritical CO2: Experimental study and modeling. Eur. J. Pharm. Sci., 106731. (2024).
Li, M. et al. Employment of artificial intelligence approach for optimizing the solubility of drug in the supercritical CO2 system. Case Stud. Therm. Eng. 57, 104326 (2024).
Shang, Y. et al. Artificial neural network hyperparameters optimization for predicting the thermal conductivity of mxene/graphene nanofluids. J. Taiwan Inst. Chem. Eng. 164, 105673 (2024).
Zhou, H. et al. Combination of group method of data handling neural network with multi-objective Gray Wolf optimizer to predict the viscosity of MWCNT-TiO2 -oil SAE50 nanofluid. Case Stud. Therm. Eng. 64, 105541 (2024).
Shadravan, S., Naji, H. R. & Bardsiri, V. K. The sailfish optimizer: A novel nature-inspired metaheuristic algorithm for solving constrained engineering optimization problems. Eng. Appl. Artif. Intell. 80, 20–34 (2019).
Srivastava, A. & Das, D. K. A sailfish optimization technique to solve combined heat and power economic dispatch problem. In 2020 IEEE Students Conference on Engineering & Systems (SCES). (IEEE, 2020).
Schapire, R. E. The strength of weak learnability. Mach. Learn. 5(2), 197–227 (1990).
Freund, Y. & Schapire, R. E. Experiments with a new boosting algorithm. In icml. ( Citeseer, 1996).
Grbić, R., Kurtagić, D. & Slišković, D. Stream water temperature prediction based on Gaussian process regression. Expert Syst. Appl. 40(18), 7407–7414 (2013).
Ma, X., Xu, F. & Chen, B. Interpolation of wind pressures using Gaussian process regression. J. Wind Eng. Ind. Aerodyn. 188, 30–42 (2019).
Rasmussen, C. E. Gaussian processes in machine learning. In Summer School on Machine Learning (Springer, 2003).
Song, H. et al. Advancing nanomedicine production via green method: modeling and simulation of pharmaceutical solubility at different temperatures and pressures. J. Mol. Liq. 411, 125806 (2024).
Williams, P. M. Bayesian regularization and pruning using a Laplace prior. Neural Comput. 7(1), 117–143 (1995).
Kruschke, J. K. Bayesian data analysis. Wiley Interdisciplinary Reviews: Cogn. Sci. 1(5), 658–676 (2010).
Kudraszow, N. L. & Vieu, P. Uniform consistency of kNN regressors for functional variables. Stat. Probab. Lett. 83(8), 1863–1870 (2013).
Cover, T. Estimation by the nearest neighbor rule. IEEE Trans. Inf. Theory. 14(1), 50–55 (1968).
Chen, C. R. & Three Kartini, U. K-nearest neighbor neural network models for very short-term global solar irradiance forecasting based on meteorological data. Energies. 10(2), 186 (2017).
Li, M. et al. Optimization of drug solubility inside the supercritical CO2 system via numerical simulation based on artificial intelligence approach. Sci. Rep. 14(1), 22779 (2024).
Almehizia, A. A. et al. Numerical optimization of drug solubility inside the supercritical carbon dioxide system using different machine learning models. J. Mol. Liq. 392, 123466 (2023).
Meng, D. & Liu, Z. Machine learning aided pharmaceutical engineering: model development and validation for estimation of drug solubility in green solvent. J. Mol. Liq. 392, 123286 (2023).
Aldawsari, M. F., Mahdi, W. A. & Alamoudi, J. A. Data-driven models and comparison for correlation of pharmaceutical solubility in supercritical solvent based on pressure and temperature as inputs. Case Stud. Therm. Eng. 49, 103236 (2023).
Acknowledgements
The authors extend their appreciation to Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia for funding this work under researcher supporting project number (PNURSP2025R205).
Funding
This work was supported by Princess Nourah bint Abdulrahman University researchers supporting project number (PNURSP2025R205), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alotaibi, H.F., Hassan, W.H., Al-Nussairi, A.K.J. et al. Computational machine learning estimation of digitoxin solubility in supercritical solvent at different temperatures utilizing ensemble methods. Sci Rep 15, 29248 (2025). https://doi.org/10.1038/s41598-025-15049-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-15049-x