抽象的
大型基因组数据集的可用性改变了微生物学研究局势。分析此类数据需要计算要求的分析,新方法来自不同的数据分析理念。机器学习和统计推断具有重叠的知识发现目标和方法。但是,机器学习的重点是优化预测,而统计推论则集中在理解与变量相关的过程中。在这篇综述中,我们概述了来自微生物基因组学的示例,概述了不同的愿望,戒律和产生的方法。强调互补性,我们认为机器学习和统计数据的结合和综合具有大数据时代的病原体研究的潜力。
背景
技术和数据生成的进步推动了微生物学的大数据革命,研究通常分析了数千个整个基因组序列。以不断增加的量,多样性和速度产生的数据集带来了巨大的机会以及独特的分析挑战。受到高通量低成本DNA测序的深入理解和驱动的启发,现在有大量的细菌物种的基因组库接近100万基因组[1]。实现这些资源的潜力需要扩展常规统计方法,这些方法面临高维数据面临挑战,需要简化和近似[2]。这是自相矛盾的,因为现代资源的庞大信息内容应该使人们更容易收集有关进化起源,传播动态和表型多样性的遗传基础的生物学见解。机器学习(ML)方法提供了潜在的解决方案,因为它们可以处理非常大而异构的数据集[3]。ML是一项多学科追求,在统计和计算机科学上大大借鉴。利用数据基于这两个努力的数据的定量方法,但是就本次审查而言,我们可以与以下区别合作:统计推断是一种提高对世界的科学理解的工具,而ML是工程自动解决方案的工具,用于预测,模拟和模式识别。
ML在生成人工智能(AI)方面取得了突破,包括自然语言,图像和音频创建[4]。在生物科学中,ML超过了对3D蛋白结构的预测的人类工程解决方案[5],将纳米孔电位转换为DNA基呼叫[6],并从大蛋白和宏基因组数据库中发现抗菌肽[7,,,,8]。在微生物种群基因组学中,当代的大数据全基因组方法通常结合统计推断和ML,以回答与传染病的进化和流行病学有关的各种问题[9]。这些包括预测未来事件(例如爆发),了解变量的影响(例如,毒力和抗药性基因),并发现数据模式(例如,感染风险中的共同点)。通常,这项工作的最佳工具不清楚或模棱两可。在这里,我们通过汇总方法和讨论示例,提供了统计推断与ML对微生物种群基因组学不同问题的适用性的一些观点。
机器学习和统计推断的原则
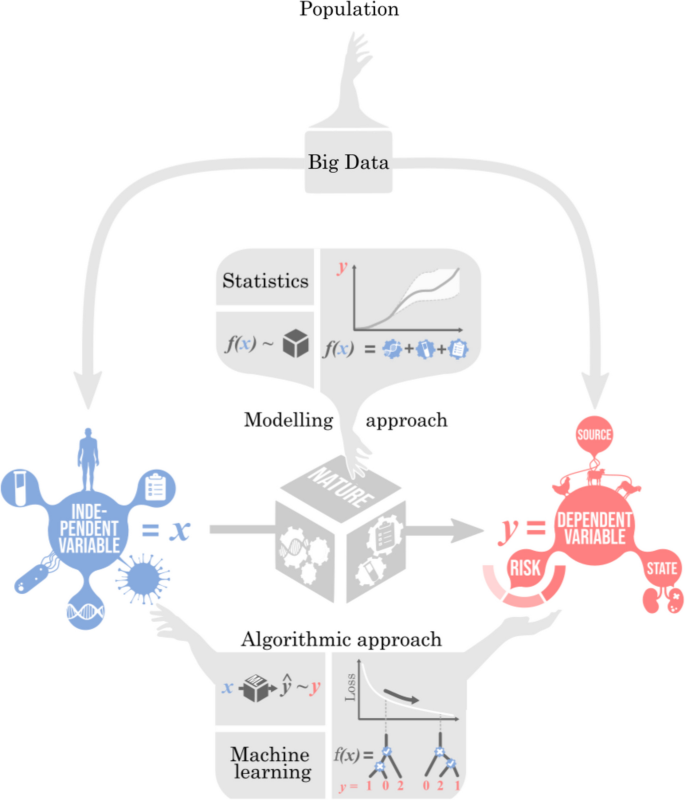
ML和统计推断是用于建模通常大型且复杂的数据的工具,这些数据已被数值编码为一个或多个变量,例如输入功能x和结果y。有关该主题的全面介绍,请参见墨菲[10,,,,11]。一个统一的概念是数据生成过程,这代表了导致手头数据的基本科学和采样过程。ML和统计数据都试图将数据生成过程近似为数学函数。从广义上讲,统计数据倾向于采用以理解潜在的愿望的模型过程,而ML采用可以忠实地复制观察到的灵活模型模式,对基础过程不可知。在建模数据生成过程的竞争方法之间有了区别:
这包含特征,也称为独立变量,预测变量或回归因子和结果,也称为因变量,标签,类或目标,因此特征的变化会导致结果的变化。关联两者的是数据生成过程或自然。统计(或更确切地说,是Breiman的二分法中的数据建模[12])旨在理解基本过程,而ML(或更确切地说,Breiman的二分法中的算法建模)旨在忠实地重现观察到的模式以实现最佳预测,例如
传统上,数据建模一直是主要的范式,尤其是在统计中,其中通过基于关于变量之间关系(确定性和随机性)之间关系的假设来衍生模型来近似数据生成过程。数据建模强调模型的解释性和建模假设的透明度。通常,考虑简约和计算负担,通常通过将现实主义与障碍性交易来选择模型复杂性。特别重点通常放在特定领域的知识和概率模型在数据建模方法中。但是,模型不必复杂,具有简单的添加线性假设为基础的工作试义(如线性回归,逻辑回归和ANOVA)。
相反,算法建模旨在为未知数据生成过程提供通用近似,而无需详细的先验知识[13]。ML的最新进展将注意力集中在算法建模上,这也包括非参数统计技术。它依靠能够在非常一般的设置中准确再现复杂数据的结构的灵活算法。这种灵活性通常需要富含参数的模型,这些模型需要大型培训数据集。因此,ML的算法开发优先计算效率。事实证明,深度神经网络特别擅长算法建模[14]。此外,ML工具包包括多种技术,其中许多都可以在PythonScikit-Learn,Pytorch和TensorFlow等软件库[15,,,,16,,,,17]。
监督与无监督的学习
在监督学习,数学函数模拟代表变量之间的关系功能x和结果y,通常是为了解释或预测y按照x。经常y低维;它可以是二进制文件,例如,描述事件是否发生,分类,描述了几种可能的结果之一或连续的结果之一。相比之下,x通常是高维的,由许多可能影响或预测感兴趣结果的投入组成。在微生物种群基因组学中,y通常是一种表型,例如药物敏感性,并且x可以代表基因组序列。基因组通常在数值上进行编码,以进行如下所述的分析。监督学习包括熟悉的方法,例如分类和回归用于建模基因型与表型关系(例如,[18,,,,19])。在无监督的学习,数学功能对数据中的关系建模x,通常以揭示隐藏的结构或模拟新数据。在最近的方法中,例如大型语言模型(LLM)x代表数字编码的文本,编号为数亿个单词[20]。在微生物基因组学中,无监督学习的重要应用是遗传聚类的检测(例如[[[21,,,,22,,,,23,,,,24])。
基因组序列数据的功能工程
在分析基因组数据之前,必须将分子序列编码为特征,或数字向量。通常,特征是根据遗传变异来定义的。两个或多个基因组之间的序列部分不同。特征通常相对于参考基因组定义。例如,单核苷酸多态性(SNP),可以编码为代表参考等位基因(例如,数量0)或非参考等位基因(1)的二元矢量的元素。如果存在多个非参考等位基因,则第二和第三个非参考等位基因由其他二进制向量表示,因此单个SNP生成多个特征,称为虚拟变量或者单速编码。同样地,等位基因在特定基因座,可以用二进制向量来表示每个基因组中每个非参考序列的存在(1)或不存在(0)。如果有k等位基因位于一个基因座,这会产生k1个功能。对于辅助基因,存在或不存在在整个基因座中,可以编码为二进制向量。
无参考的方法也很受欢迎。基因组组件或蛋白质序列可以切成寡核苷酸或寡肽的短而重叠的窗户k-Mers,在哪里k表示序列长度。每个人的存在或不存在k - 每个基因组中的子可以编码为二进制向量。很短k-Mers(k<5)关于核苷酸组成的信息,而k - 50范围内的船员可以捕获SNP,Indels和基因存在或不存在的基因座特异性变化。如果k更长,k-MERS变得稀有或独特,因此对于推论或预测而言有用。更高级的用途k-Mers在保留其生物学含义的同时减少功能的数量,例如通过合并k - 始终(或有时)共享相同(或相似)存在模式与基因组不存在的模式Unitigs(或者嵌入)编码为二进制(或连续)向量(例如,24,,,,25])。生物问题和分析目标根据生物学问题牢固地构建分析的目标有助于缩小适当的ML或统计推断工具。
生物学问题映射到分析目标,包括(i)数据探索,(ii)预测,(iii)参数估计和(iv)假设检验。
在数据探索,目的通常是熟悉,可视化或假设产生。这些目标是开放式的,但是它们具有识别或传达数据重要特征或不丢失数据重要方面的重要特征的共同主题。通常,分析目标可以通过损失功能这是约束或最小化的。在比较ML和统计方法时,考虑损失功能有助于。
在预言,目的是通过利用观察到的数据中的模式来预测,插入,分类或模拟新的,未观察或故意掩盖的数据,同时最大程度地减少预测错误:真相与预测之间的差异。预测的常见损失函数包括连续结果的平方误差或离散结果的0 1个错误分类错误,其中1表示错误分类,0表示正确的分类[26,,,,27]。
在参数估计,目的是精确量化假定描述数据生成过程的数学模型的参数。估计的常见损失函数包括误差,绝对误差和平方错误。最后,在假设检验,目的是得出定性的结论,例如,变量会影响结果。在这里,假阳性通常被编码为0 1损耗函数,该函数指示零假设是否已被错误拒绝(1)(0)。
比较性能
可以通过在观察到的数据点上平均损失函数来比较ML和统计方法的性能(经验风险),或跨先前的分布(贝叶斯风险),或跨理论重新制定数据生成过程(频繁的风险)。经验风险很方便,但需要一个地面真相,因此最适用于预测,在该预测中可以将预测直接与被掩盖或设置的观察到的数据进行比较以衡量预测准确性。ML为预测提供了丰富的灵活算法工具箱,这通常可以帮助分析师实现与单独使用传统统计方法更小的经验风险。
统计方法在无法获得地面真理时会有所帮助,例如,在估计参数并测试有关未观察到过程的假设时。最大似然估计和似然比测试是广泛使用的经典方法,可最大程度地减少或限制频繁的风险,例如平方误差(用于估计)和家庭误差率(用于假设检验)。这些保证需要遵守技术假设,例如大型样本量和(用于假设检验)模型的嵌套。当我们愿意对未知参数的可能值进行事先假设时,贝叶斯推断对于参数估计和假设检验有用,因为它可以最大程度地减少或约束贝叶斯损失函数,例如均方误差(用于预测或估计)和假发现率(对于假设检验)。它不依赖于大型样本量之类的假设,但是贝叶斯的方法在计算上可能是强化的。
拟合模型
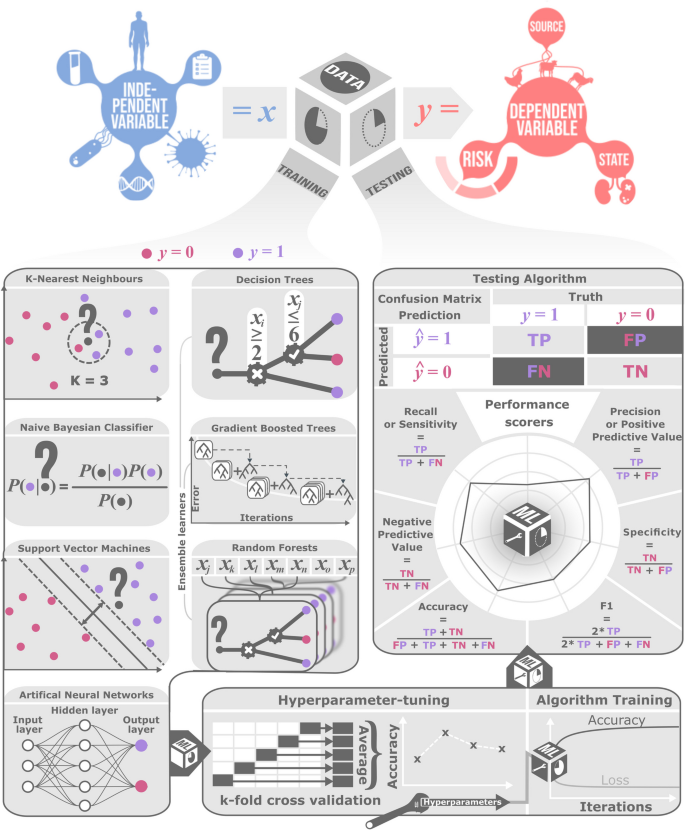
数据通常分为培训和测试当有地面真相时,设置可以将经验风险最小化(在培训期间)并进行测量(在测试期间)。使用培训数据优化参数,然后使用测试数据评估最终性能。这个想法是要获得对性能的独立,公正的估计,但这可能会因培训和测试数据之间的依赖而破坏。有时ML模型需要在训练过程中难以适应的超参数,因此验证设置用于使用网格搜索来优化它们(图 2)。交叉验证是一种流行的技术,用于平均分解数据的不同方式[28]。在古典和贝叶斯统计中,为了进行估计和假设测试,通常将整个数据用于适合模型,因为贝叶斯风险或频繁的风险可以从理论上优化。这可以更有效地使用数据。
在ML和统计推断中,特别是在参数富含参数或数据限制的设置中,过度合身风险噪声参数估计值和对其他数据的可推广性差[29,,,,30]。为了减轻过度拟合,练习是很常见的正则化,其中参数值以某种方式受到约束。正则化的例子包括受到惩罚的可能性和贝叶斯先验。集合方法,例如在随机森林中聚集的自举,并在梯度增强的树木中提升,也通过优化伪复制数据的性能来减少过度拟合。相比之下,辍学在人工神经网络中,通过优化随机修剪的网络的性能来避免过度拟合以建立弹性,并避免在训练过程中对神经元的过度特殊化。通常可以调节ML中的训练算法以减少过度拟合,通过修改称为The的调谐参数学习率,并制定称为称为的策略早期停止规则。对过度拟合的担忧必须与模型不佳或不合身的过度纠正,这是一种称为平衡偏见变化权衡。
机器学习分类器在微生物基因组学中常见
在分类,挑战是预测或解释结果变量,y,使用功能中的信息,一个固定数量值(或标签)之一的分类变量(或类),x。通常,算法具有通过优化训练数据集中的精度来校准的参数。在微生物基因组分析中使用了几种常见的ML分类器,其复杂程度不同。最早的分类算法是k-near最邻居。在这里,推断的班级是在k培训数据最接近的数据点x,从某种意义上说。这需要一个距离度量[31,,,,32]。应用包括从DNA序列中预测基因功能和表型[33,,,,34]。另一个相对简单的方法是高度可扩展的NAN -VE贝叶斯方法,使用贝叶斯定理分配类,假设特征之间具有独立性。在这里,推断的类是后验概率最高的类[35,,,,36,,,,37]。假定有条件的可能性,假定必须学习统计分布(例如高斯,伯努利)。应用包括疾病诊断[38]和基于序列的基因组分类法,元基因组[39]和水平转移的基因[40]。
有几种更复杂的方法,包括支持向量机,决策树和人工神经网络。支持向量机基于内核提供了一种灵活的分类方法,该方法测量了数据点之间的特征的相似性。非线性内核有助于在图像分析等困难问题中进行分类。结果可能对调整参数敏感[41,,,,42,,,,43]。应用包括检测水平基因转移[44],从基因组序列中预测分子表型[45,,,,46]和分类主机特异性[47]。
决策树可以将用于鉴定物种的生物学现场指南中的键进行比较。在这里,决策树代表了使用功能分配标签或类的规则的层次结构序列。使用启发式贪婪算法对规则进行了训练,并修剪以减轻过度拟合。易于解释的单个决策树通常被使用合奏来提高准确性并降低噪音[48,,,,49]。知名随机森林是一种合奏方法,在训练构建许多决策树时,重复对功能和数据点进行反复进行(自举)。使用跨树的最常见分类(汇总),它提高了准确性[50,,,,51,,,,52]。应用包括预测致病性,疾病状况,抗菌素抵抗,基因组含量和宿主特异性[53,,,,54,,,,55,,,,56,,,,57,,,,58,,,,59,,,,60,,,,61,,,,62]。梯度树的增强是逐步种植决策树森林的另一种合奏方法,最后一棵树训练以改善上一步的预测,通过损失功能进行评估[63,,,,64,,,,65]。应用包括预测相关基因序列的pH偏好和抗菌抗性[66,,,,67]。
最后,受神经科学的启发,人工神经网络(ANNS)已经成为微生物基因组学中流行的ML方法。ANN包括简单功能(人造神经元)的有向图(网络)。ANN的体系结构各不相同,但通常将神经元组织成观察到的(输入和输出)层和一个或多个隐藏层[68]。沟通发生在安的层之间[69]。深度学习使用具有多个隐藏层的ANN,该ANN生产具有大量信息处理能力的复杂而灵活的模型[70,,,,71]。大数据可用性,GPU(图形处理单元)和理论创新的进步已使参数丰富的ANN有效地拟合。应用包括从DNA序列中识别物种,菌株和基因功能[72,,,,73,,,,74,,,,75]。ANN的表现很好,部分原因是它们通过近似任意连续关系充当通用函数近似器,给定足够的隐藏神经元[76],部分是因为拟合技术被认为施加正则化(例如77])。注意机制启用一些安慰剂,特别是变压器,动态加权基于上下文的输入元素的影响,而不是依靠固定的连接模式[78]。这使网络可以选择性地关注输入的最相关部分,无论其位置如何。注意对于分子序列或三维蛋白质结构的依赖性很有用,在传统体系结构中难以传播远程信息。注意机制允许每个输入元素并行直接考虑所有其他元件,从而避免了重要但遥远的信号的稀释。注意引起了生成AI的突破[79],抗生素预测[7,,,,8]和蛋白质结构预测[5]。
机器学习和统计的优势和劣势
对生物学问题的明确陈述通过确定最小化的损失类型来为分析目标提供信息。最小化估计误差与预测误差与误报的最小化指导了方法的选择。旨在理解基本过程因果关系的数据分析可以通过统计推断为有因果关系提供,因为它将最大程度地减少与估计和假设检验相关的(贝叶斯或频繁的)风险。ML可以更好地为旨在优化模型解决问题的数据分析,因为它可以在可用的地面真理时最大程度地减少预测的(经验)风险[29,,,,80]。主要的统计范式强调了这样的原则简约和解释性,而复杂的ML算法可以比统计中常见的简单模型产生出色的性能。诸如XOR问题之类的经典监督学习示例来说明这一点,在XOR问题中,输出不是输入数据的线性函数。
开箱即用,许多ML方法处理共线性,,,,非线性, 和互动比传统的统计方法(例如回归)更好。一位经验丰富的统计学家可能会采用正规化来应对不可靠的参数估计值和由密切相关或共线特征引起的高不确定性,但正则化在许多ML算法中内置为标准化。特征和结果之间的非线性关系,并且功能之间的相互作用也可以统计地建模,但是这需要数据分析师部分的一些复杂性和手动干预,而许多ML算法旨在自动建模这些现象。ML算法通常可以在数千个功能中优先考虑,从而使用户可以采用不可知论方法来选择功能。但是,复杂的ML的成本是其工作和参数与解释的透明度的模型[81],经常被称为黑匣子[19]。
机器自动化的强劲表现和模型不可知论的优势取消了人们对人类问责制的重要性数据质量问题;这被称为自动化偏见。有偏采样和批处理效果通过产生可能具有误导性或易于普遍的结论来为ML和统计推断造成问题(请参阅数据质量和询问结果)。此外,有关解释性,,,,平等, 和问责制在许多情况下很重要,尤其是医疗保健[82]。因此,在特定的损失函数意义上的模型表现之间存在的权衡取舍,而在社会上更广泛的效用可能会改变优先ML与统计推断之间的平衡。ML vs统计和数据建模与算法建模二分法都回想起更基本的区别演绎(基于逻辑)vs感应(基于观察的)科学推断。从根本上讲经验ML建模的方法是数据驱动的和渴望数据的方法,解释了其对大数据和对偏见数据集的敏感性的依赖,但它的较高灵活性可以更紧密地拟合数据。
数据质量和询问结果
格言 - 垃圾,垃圾是ML和统计数据中的一种真实性:适当的数据准备和质量检查(QC)对于任何分析都是必不可少的。研究人员必须采取策略来诊断数据质量分析前后问题。
第一步,必须了解出处数据,其局限性以及是否足以满足分析目标。接下来,必须使用包括摘要统计信息和视觉效果在内的方法对数据进行质量检查,以诊断诸如数据输入错误,异常值,缺失值和特殊值等问题。必须正确编码数据,尤其是缺少或特殊值,以确保ML或统计算法适当处理它们。一个插补步骤,为了预测缺失值。除QC外,数据探索对于假设产生并选择了合理假设的合适模型很有价值。
在合并可能在不同地方,不同时间,不同过程或出于不同目的中收集的数据集之前,重要的是要考虑如何受到分析的影响异质性数据集之间的系统差异。例如,可能有未得到的混淆者这之间有所不同。跨数据集结果的系统差异使分析特别容易受到所谓批处理效果。有时,通过将批次标签作为特征来控制异质性。一种更强大但效率较低的方法是荟萃分析,其中分别分析数据集并比较结果后的结果,并在适当的情况下合并。通常,这整齐地适合培训,测试和验证,尤其是因为样本外预测比拆分单个数据集更强大的通用性指标。
经过分析后,必须再次询问数据,以了解信息信号的来自何处并诊断未解决的质量控制问题或实施错误。健康怀疑论,特别是对于令人惊讶的结果,很重要,并且考虑到以下问题:(i)结果如何与文学?(ii)结果对分析假设的鲁棒性吗?针对更简单的方法进行基准测试可以在这里有所帮助。除非可以解释数据中的信号,例如使用可视化或可解释的AI,可能很难说服同伴。实验验证和复制在独立的数据集中,通常需要建立信誉,并重复另一种truism,``特殊的主张都需要非凡的证据。”
ML和统计在微生物基因组中的应用
在本节中,我们考虑了ML和统计数据在微生物基因组学中的应用,并在三个例子的背景下讨论了竞争方法的相对优势:人畜共患细菌中的源归因,全基因组全基因组抗性抗性研究的研究,并预测了基因组序列的抗菌抗性抗菌抗性。
示例1:弯曲杆菌中的来源归因
#PREDICTION #Classification #supervise_learning #machine_learning。
特征 (x):基因组序列。结果(y):原产地。
鉴定细菌感染的原始群体对一系列病原体具有实际应用沙门氏菌,,,,大肠杆菌, 和弯曲杆菌。人与人的传播弯曲杆菌这是人类胃炎的常见原因,很少见,大多数情况是由于食用受污染的食物而引起的。弯曲杆菌通常将鸟类和哺乳动物的胆量殖民,包括用于肉类和家禽的动物,并在环境水中发现。因此,每个人类案例都被认为源自其中一个源储存库,并且预测或属性源是有用的。来源归因有助于通过告知破坏传输链的努力来防止未来的人类案件。
DNA测序已被利用用于源归因弯曲杆菌使用各种工具。数据通常包含DNA序列弯曲杆菌从人类感染病例中分离出来,并从动物和环境库中进行比较。早期方法采用统计流行病学模型,使用应变水平的名称来排除传播(例如[[83,,,,84])。后来,统计模型以人群遗传学为基础,结构和Isource应用,应用,利用DNA中的更多信息[85,,,,86]。但是,可以将源归因作为直接的ML问题提出,其中分析目标是最大程度地减少错误分类误差。DNA序列弯曲杆菌直接从源种群采样可用于训练具有已知标签(例如牛,绵羊,猪,鸡,环境水)的分类器。可以使用交叉验证测试分类器精度。然后可以从DNA序列预测每个人类病例的原始群体。事实证明,ML分类器比应用于多层次序列键入(71%vs 64%)的已建立的统计方法更快,准确性约11%,并且很容易被推广到整个基因组测序(WGS)数据的分析,允许准确的33%增长(85%vs 64%)[[85%)[[85%)[[64%)[58,,,,59]。随机森林[51]和xgboost [87]产生了最大的进步。在这种情况下,ML成功的关键是大数据的可用性,其中包括数千个具有高度复制的整个基因组:从感兴趣的来源种群中采样了5799个基因组,以及人类感染中的15,988个基因组。
示例2:全基因组抗菌素耐药性研究
#hypothesis_testing #parameter_estimation #regression #statistics。
特征 (x):基因组序列。结果(y):抗菌素耐药性或灵敏度。
A major aim of biology in the twenty-first century is to unravel the genetic architecture of phenotypic diversity within species [88]。In microbiology, there is particular interest in traits that affect the outcome of human colonization and infection, like virulence (the frequency or severity of disease) and antimicrobial resistance (AMR).Early approaches to such questions studied candidate genes, for example using PCR to test for differences in the frequency of genetic markers between cases and controls (e.g., [89])。With the advent of technologies like genotyping arrays and, later, whole-genome sequencing, the accepted approach to such questions has been to scan the genome for evidence of associations between allelic differences and phenotypic differences.So-called genome-wide association studies (GWAS) address concerns that candidate gene approaches are vulnerable to selection and reporting bias, and struggle to control artefactual associations caused by population stratification of phenotypes, for example when phenotypes differ between strains [90,,,,91,,,,92]。
GWAS is motivated by a desire to learn about the causal process underlying the data, and pains are taken to avoid artefactual signals of association, while recognizing that observational studies cannot prove causality (see, e.g., [93,,,,94])。This is a statistical inference problem in which the parameters of a relatively simple and readily interrogated general linear model are interpreted to identify genetic variants responsible for observable phenotypic diversity.Special emphasis is placed on limiting the expected losses caused by false positive associations.In bacteria, GWAS have been applied to a range of traits and species (e.g., [54,,,,95,,,,96,,,,97,,,,98,,,,99])。While ML approaches have been applied to this problem, and while informative for data exploration and hypothesis generation, particularly in expert hands [100], ML approaches only return “high-leverage†genes or genetic variants that help predict the outcome.Out-of-the-box they neither test nor quantify the evidence for the hypothesis that these variants directly influence the outcome.Nor do they offer theoretical or empirical tools for easily controlling family-wise error or false discovery rates across loci.Statistical approaches address these foundational issues, and mapping of genes underlying AMR has proved particularly fruitful (e.g., [101,,,,102]), presumably because mechanisms of genetic resistance are often direct, almost deterministic.GWAS depends on big data to find signals of association, but interpretation of those signals relies on explicit modelling assumptions, and not on training a general-purpose algorithm using datasets of many known genotype-to-phenotype associations, which as yet do not exist.
Example 3: predicting antimicrobial resistance from genome sequences
#prediction #classification #supervised_learning #interpretable_machine_learning.
特征 (x): genome sequences.Outcomes (y): antimicrobial resistance or sensitivity.
Related to the problem of inferring which genes confer antimicrobial resistance is the problem of predicting antimicrobial resistance from an individual bacterial genome.Modernizing microbiological diagnostics in clinical practice has been a major focus of research over the last 15Â years, with aspirations to replace a battery of phenotypic tests with a streamlined WGS and phenotype prediction pipeline [103]。WGS has become routine in some healthcare settings, particularly for organisms that are challenging to test in the laboratory, like the slow-growing and high biosafety level pathogenMycobacterium tuberculosis[104,,,,105]。
The statistical models used for GWAS could be turned to prediction, but the superior flexibility of ML algorithms to fit data more closely make them a natural choice for predicting AMR (e.g., [100,,,,106,,,,107,,,,108,,,,109,,,,110])。In this setting, the analysis goal is to minimize prediction error, which can be quantified empirically because a ground truth is available.Large datasets have been generated comprising WGS and traditional AMR phenotyping assays and based on these, automated predictions with high accuracy have been achieved—in some cases exceeding the standards required of traditional laboratory diagnostics [111,,,,112]—confirming the excellent performance of ML algorithms for general-purpose prediction.
ML performance in AMR prediction has established it as an important tool for predicting all manner of bacterial phenotypes from WGS data.However, there is a question of accountability: in a medical setting, decision-taking responsibility lies with the clinical microbiologist.Therefore the ML algorithm needs to present the evidence for its prediction transparently for interpretation by the domain-specific expert.Scenarios like this create a need forexplainable AIthat goes significantly beyond outputting coefficients for predictive features, which may be mere confounders, rather than biologically causal genetic variants, particularly in the presence of population stratification [113]。Approaches to explainable AI includeattribution algorithms, which may impose post hoc linearization of the predictions (e.g., [114,,,,115])。This leads back to simpler, more transparent data models resembling additive or linear models.或者,ablation algorithmssystematically drop features-of-interest from the model to assess their impact on performance [116]。Consequently, even when pursuing prediction via complex ML, efforts to interpret prediction may resemble more traditional statistical analysis in which high importance is attached to understanding in a causal way the conclusions and interpretation of the data.
统计与机器学习:正确的工作工具
The boundary between ML and statistics is blurred, with cross-over methods like the elastic net, bootstrap, non-parametric statistics, and Bayesian-inspired approaches.The labels “machine learning†and “statistics†are typically less useful than a clear definition of the analysis goals—prediction, exploratory data analysis, parameter estimation, hypothesis testing—which in turn are framed by the biological questions.Where a project has multiple goals, such as prediction and hypothesis testing, it is reasonable to apply different analysis approaches to the same data.However, as example 3 illustrates, even when a task clearly fits the goal of prediction, the choice of method is influenced by context-specific considerations, notably explainability and accountability.Frequently in scientific applications, there is an emphasis on understanding and interpreting the data generating process, and this may tip the balance away from ML and towards statistical inference.Interrogating results in real data analysis, detecting data quality issues like batch effects, explaining which signals drive the results, controlling for confounding factors, and understanding the limits to generalizability, are essential to the integrity of scientific outputs.Developing strategies to check scientific results is a key step towards scientific independence that allows a researcher to take responsibility for final conclusions.The risk of automation bias, in which responsibility for final conclusions is delegated to opaque algorithms, and abdication of critical thinking, are rightly of concern.
结论和未来的方向
We are currently in a period of exploration, as ML and AI are increasingly applied to diverse questions like “what is the genetic architecture of virulence,†“why do dangerous pathogens emerge,†and “how do we fight the spread of antimicrobial resistanceâ€?In allied fields, we have seen transformative innovations ranging from the prediction of 3D molecular structure [5] to antimicrobial peptide discovery [7,,,,8] and, looking ahead, the design of novel proteins and molecular systems based on free text (e.g., [117,,,,118,,,,119,,,,120])。In microbial population genomics, we anticipate that ML will continue to play a leading role, both by improving on previous approaches, and by opening new avenues of research and understanding.If, in the years to come, there were to be a final analysis of the role of ML and AI in microbial genomics, no doubt it would re-emphasize the enduring importance of deductive statistical thinking, currently less fashionable as the new opportunities presented by ML take precedence.Statistics provides a foundation for scientific thought, clarifying concepts like study design, randomization, replication, control, batch effects, mediation and confounding, causation, and correlation.Scientific progress is a continual process, so there will be no final analysis.Instead, we expect a gradual assimilation of recent developments in AI/ML together with well-established statistical approaches into a new and emerging field of Data Science.
Glossary 1: Terms in statistical inference and machine learning, generated by ChatGPT-4o and manually curated
Attribution algorithms
Methods that assign importance scores to input features by estimating their contribution to a model’s prediction, often using gradients, perturbations, or local surrogate models (e.g., SHAP, LIME).
Ablation algorithms
Techniques that assess feature importance by systematically removing or masking input features and measuring the resulting impact on model performance or predictions.
Automation bias
The tendency for humans to over-rely on automated systems, such as ML models, even when they may be incorrect.
Batch effects
Non-biological variations introduced into data during different processing times, instruments, or sample batches, which can confound results.
Bias-variance trade-off
A fundamental concept that describes the balance between underfitting and over-fitting.High bias models are too simple and may miss patterns (underfitting), while high variance models are too complex and may capture noise as if it were signal (over-fitting).Optimal performance is achieved by balancing these two sources of error.
Biased sampling
Occurs when the sample used for training or testing a model is not representative of the overall population-of-interest, leading to biased, misleading, or non-generalizable results.
黑匣子
A term used to describe models (such as deep neural networks) that are complex and difficult to interpret, where the internal workings are not easily understood.
Computational efficiency
Refers to the amount of computational resources (time and memory) required to train and use a model.More efficient models can handle larger datasets or run faster.
共线性
Collinearity occurs when two or more features are highly correlated, meaning they share a linear relationship.This makes it difficult to estimate the unique contribution of each predictor, leading to unreliable estimation with high uncertainty.
Cross-validation
A technique for assessing how well a model generalizes to unseen data by partitioning the dataset into multiple subsets and training/testing the model on different combinations of these subsets.
Data generating process
The scientific and sampling mechanisms by which the observed data in a study were produced.
Data quality
Refers to the accuracy, completeness, and reliability of data, which directly affects the performance of statistical inference and ML.
Data vs algorithmic modelling
In Breiman’s dichotomy,data modellingfocuses on building models that capture the essence of the underlying data generating process, whereasalgorithmic modellingfocuses on flexible prediction algorithms that exploit the structure in the observed data, without making assumptions about the underlying data generating process.
深度学习
A subset of ML that involves neural networks with many layers (deep architectures) used to model complex patterns in data, particularly useful for image, speech, and sequence tasks.
Deductive vs inductive reasoning
Deductive reasoningdraws specific conclusions from general principles or theories, whereas归纳推理infers general patterns or rules from specific observations or data.
Domain-specific knowledge
Expert knowledge about the particular field or domain of application (e.g., genomics) that helps guide model development and interpretation of results.
Dropouts
A regularization technique commonly used in neural networks where random units (artificial neurons) are “dropped†or ignored during training to prevent over-fitting.
Early stopping rules
A technique used to stop training a model once its performance on a validation set starts to degrade, preventing over-fitting.
经验
Based on observation or experimentation rather than theory.Empirical data is gathered from real-world experiments or observations.
Ensemble methods
ML methods that combine the predictions of multiple models to improve accuracy and robustness.Common examples include Random Forest and Gradient Boosting.
解释性
The ability to interpret and understand how a ML model makes decisions, particularly in complex or high-dimensional models.
False discovery rate (FDR)
The expected proportion of false positives among all rejected null hypotheses in multiple hypothesis testing.
Family-wise error rate (FWER)
The probability of making one or more false positive errors when performing multiple hypothesis tests.
特征
Individual measurable properties or characteristics of the data used to train a model;they are analogous to independent variables in traditional statistical terminology, representing the inputs used to predict or explain an outcome.
Hypothesis test
A statistical method used to determine whether there is enough evidence to reject a null hypothesis, usually based on the comparison of a test statistic to a critical value.
互动
Interactions occur when the effect of one feature on the outcome depends on, or is modified by, the value of another feature.
Interpretable machine learning
A branch of ML focused on developing models that provide human-understandable explanations for their predictions.
Interpretability, equality, and accountability
Important ethical considerations in ML that refer to the clarity of model outputs (interpretability), fairness across different groups (平等), and responsibility for decisions made by models (问责制)。
Learning rate
A hyper-parameter that controls how much a ML model’s weights are updated with respect to the gradient of the loss function during training.
损失功能
Quantifies the (lack of) quality of a model’s performance relative to the biological aims.It guides the optimization process during training.Examples include the mean squared error, calculated between a prediction or estimate and the truth, and 0–1 loss, where a value of 1 indicates a misclassification error or a false positive.Usually it is风险, rather than loss, that is minimized.
Maximum likelihood estimate (MLE)
A method of estimating the parameters of a statistical model by maximizing the likelihood function, which measures how likely it is to observe the given data under different parameter values.
最大限度a posterioriestimate (MAP)
An estimation method that incorporates prior knowledge or beliefs about the parameters in addition to the likelihood of the data, often used in Bayesian statistics.
Non-linearity
Non-linearity refers to relationships between variables that cannot be adequately captured by a straight line.In a non-linear relationship, changes in one variable do not simply lead to proportional changes in another.
结果
The target variables that a model aims to predict or explain;they are analogous to dependent variables in traditional statistical terminology, representing the outputs that depend on the input features.
Over-fitting
Occurs when a model learns not only the underlying pattern in the training data but also the noise, leading to poor performance on new, unseen data.
Parsimony
A principle that prefers simpler models over more complex ones when both explain the data equally well, often used interchangeably with Occam’s Razor.
Probabilistic models
Models that incorporate uncertainty by assigning probabilities to different outcomes, useful for reasoning about uncertainty in data.
Python
A high-level programming language widely used in data science and ML due to its simplicity, extensive libraries and strong community support.
Regression vs classification回归
models continuous outcomes, while分类models categorical outcomes, in both cases fitting observed or predicting new outcomes based on the features of other input data.正则化
A technique used to prevent over-fitting by incorporating a penalty on the parameter values into the loss function (e.g., L1, L2 regularization).
风险
The expected value of a
损失功能, defined as an arithmetic mean over (i) observed datapoints (empirical risk), (ii) a prior distribution (Bayes risk), or (iii) hypothetical repetitions of the data generating process (frequentist risk)。Usually it is risk, not loss, that can be minimized by model fitting.
Supervised vs unsupervised learning
在监督学习, a statistical or ML algorithm is trained on labelled outcome data, whereas in无监督的学习, the algorithm learns from unlabelled data, discovering patterns without explicit outcomes.
Training, testing, and validation
一个训练集comprises data used to fit or train a model.一个validation setis a separate subset of data used to tune model parameters and assess performance during training, where necessary.这test setis another, separate set of data used to evaluate the model’s performance after training is complete.
Data availability
在当前研究中没有生成或分析数据集。
参考
Blackwell GA, Hunt M, Malone KM, Lima L, Horesh G, Alako BTF, et al.Exploring bacterial diversity via a curated and searchable snapshot of archived DNA sequences.Plos Biol。2021;19:e3001421 (Hanage WP, editor.).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wong ZSY, Zhou J, Zhang Q. Artificial intelligence for infectious disease big data analytics.感染DIS健康。2019;24:44–8.
文章一个 PubMed一个 Google Scholar一个
Ow GS, Tang Z, Kuznetsov VA.Big data and computational biology strategy for personalized prognosis.Oncotarget.2016;7:40200–20.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Bommasani R, Hudson DA, Adeli E, Altman R, Arora S, von Arx S, et al.On the Opportunities and Risks of Foundation Models.arxiv;2021 Available from:https://arxiv.org/abs/2108.07258. [cited 2025 Sept 2].
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al.Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature.2024;630:493–500.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Pagès-Gallego M, De Ridder J. Comprehensive benchmark and architectural analysis of deep learning models for nanopore sequencing basecalling.基因组生物。2023;24:71.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Torres MDT, Brooks EF, Cesaro A, Sberro H, Gill MO, Nicolaou C, et al.Mining human microbiomes reveals an untapped source of peptide antibiotics.细胞。2024;187:5453-5467.e15.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Wan F, Torres MDT, Peng J, De La Fuente-Nunez C. Deep-learning-enabled antibiotic discovery through molecular de-extinction.Nat Biomed Eng。2024;8:854–71.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Iwashyna TJ, Liu V. What’s So Different about Big Data?.A Primer for Clinicians Trained to Think Epidemiologically.Annals ATS.2014;11:1130–5.
Murphy KP.Probabilistic machine learning: an introduction.Cambridge, Massachusetts: The MIT Press;2022。
Murphy KP.Probabilistic machine learning: advanced topics.Cambridge, Massachusetts: The MIT Press;2023。
Breiman L. Statistical Modeling: The Two Cultures (with comments and a rejoinder by the author).Statist Sci.2001;16.Available from:https://projecteuclid.org/journals/statistical-science/volume-16/issue-3/Statistical-Modeling--The-Two-Cultures-with-comments-and-a/10.1214/ss/1009213726.full. [cited 2025 Sept 2].
Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning.NAT方法。2018;15:233–4.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Schmidhuber J. Deep learning in neural networks: an overview.神经网。2015;61:85–117.
文章一个 PubMed一个 Google Scholar一个
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O. Scikit-learn: Machine learning in Python.J Mach Learn Learn Res。2011;12:2825–30.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al.PyTorch: An Imperative Style, High-Performance Deep Learning Library.Advances in Neural Information Processing Systems 32. Curran Associates, Inc;2019;8024–35.
TensorFlow Developers.张量。Zenodo;2024. Available from: https://zenodo.org/doi/10.5281/zenodo.12726004. [cited 2025 Sept 2].
Greene AC, Giffin KA, Greene CS, Moore JH.Adapting bioinformatics curricula for big data.简短的生物知识。2016;17:43–50.
文章一个 PubMed一个 Google Scholar一个
Wiemken TL, Kelley RR.Machine learning in epidemiology and health outcomes research.Annu Rev公共卫生。2020;41:21–36.
文章一个 PubMed一个 Google Scholar一个
Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al.Language models are few-shot learners.Adv Neural Inf Process Syst.2020;33:1877–901.
Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al.Traces of human migrations in幽门螺杆菌幽门螺杆菌人群。科学。2003;299:1582–5.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Corander J, Marttinen P. Bayesian identification of admixture events using multilocus molecular markers.Mol Ecol。2006;15:2833–43.
文章一个 PubMed一个 Google Scholar一个
Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure.核酸res。2019;47:5539–49.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lees JA, Tonkin-Hill G, Yang Z, Corander J. Mandrake: visualizing microbial population structure by embedding millions of genomes into a low-dimensional representation.Phil Trans R Soc B. 2022;377:20210237.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Jaillard M, Lima L, Tournoud M, Mahé P, Van Belkum A, Lacroix V, et al.A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events.Didelot X, editor.PLOS基因。2018;14:e1007758.
Hoffman S, Podgurski A. Big bad data: law, public health, and biomedical databases.J Law Med伦理。2013;41:56–60.
文章一个 PubMed一个 Google Scholar一个
Wang Q, Ma Y, Zhao K, Tian Y. A comprehensive survey of loss functions in machine learning.Ann Data Sci.2022;9:187–212.
文章一个 Google Scholar一个
Stone M. Cross-Validatory Choice and Assessment of Statistical Predictions.J Royal Statistic Soc Series B (Methodological. 1974;36:111–47.
Bzdok D, Krzywinski M, Altman N. Machine learning: a primer.NAT方法。2017;14:1119–20.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Bashir D, Montañez GD, Sehra S, Segura PS, Lauw J. An Information-T.CHAM:Springer International Publishing;2020;347–58.Available from:https://link.springer.com/10.1007/978-3-030-64984-5_27. [cited 2025 Sept 2].
Fix E, Hodges JL.Discriminatory analysis: Nonparametric discrimination: Consistency properties: (471672008–001).1951 Available from:https://doi.apa.org/doi/10.1037/e471672008-001. [cited 2025 Sept 2].
Cover T, Hart P. Nearest neighbor pattern classification.IEEE Trans Inform Theory.1967;13:21–7.
文章一个 Google Scholar一个
Yao Z, Ruzzo WL.A regression-based K nearest neighbor algorithm for gene function prediction from heterogeneous data.BMC生物信息学。2006;7:S11.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
MihelÄić M, Å muc T, Supek F. Patterns of diverse gene functions in genomic neighborhoods predict gene function and phenotype.Sci Rep. 2019;9:19537.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Xu S. Bayesian naïve Bayes classifiers to text classification.J Inf Sci.2018;44:48–59.
文章一个 Google Scholar一个
John GH, Langley P. Estimating Continuous Distributions in Bayesian Classifiers.arxiv;2013 Available from:https://arxiv.org/abs/1302.4964. [cited 2025 Sept 2].
Webb GI.Naïve Bayes.In: Sammut C, Webb GI, editors.Encyclopedia of Machine Learning.Boston, MA: Springer US;2011713–4.Available from:https://link.springer.com/10.1007/978-0-387-30164-8_576。[cited 2025 Sept 2].
Li F, Shen Y, Lv D, Lin J, Liu B, He F, et al.A bayesian classification model for discriminating common infectious diseases in Zhejiang province, China.药品。2020;99:e19218.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zhao Z, Cristian A, Rosen G. Keeping up with the genomes: efficient learning of our increasing knowledge of the tree of life.BMC生物信息学。2020;21:412.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sandberg R, Winberg G, Bränden C-I, Kaske A, Ernberg I, Cöster J. Capturing whole-genome characteristics in short sequences using a naïve Bayesian classifier.基因组res。2001;11:1404–9.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Ben-Hur A, Ong CS, Sonnenburg S, Schölkopf B, Rätsch G. Support vector machines and kernels for computational biology.PLoS Comput Biol.2008;4:e1000173 (Lewitter F, editor.).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
McIntyre ABR, Ounit R, Afshinnekoo E, Prill RJ, Hénaff E, Alexander N, et al.Comprehensive benchmarking and ensemble approaches for metagenomic classifiers.基因组生物。2017;18:182.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Cortes C, Vapnik V. Support-vector networks.马赫学习。1995;20:273–97.
文章一个 Google Scholar一个
Tsirigos A. A sensitive, support-vector-machine method for the detection of horizontal gene transfers in viral, archaeal and bacterial genomes.核酸res。2005;33:3699–707.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Weimann A, Mooren K, Frank J, Pope PB, Bremges A, McHardy AC.From Genomes to Phenotypes: Traitar, the Microbial Trait Analyzer.Segata N, editor.mSystems.2016;1:e00101–16.
Belman S, Pesonen H, Croucher NJ, Bentley SD, Corander J. Estimating Between Country Migration in Pneumococcal Populations.Epidemiology;2023。可从以下方式获得:http://medrxiv.org/lookup/doi/10.1101/2023.11.15.23298520。[cited 2025 Sept 2].
Lupolova N, Dallman TJ, Holden NJ, Gally DL.Patchy promiscuity: machine learning applied to predict the host specificity of Salmonella enterica and Escherichia coli.Microbial Genomics.2017; 3。Available from:https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000135. [cited 2025 Sept 2].
Quinlan JR.Induction of decision trees.马赫学习。1986;1:81–106.
文章一个 Google Scholar一个
Li M, Xu H, Deng Y. Evidential decision tree based on belief entropy.熵。2019;21:897.
文章一个 PubMed Central一个 Google Scholar一个
Schrider DR, Kern AD.Supervised machine learning for population genetics: a new paradigm.趋势基因。2018;34:301–12.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Breiman L. Random forests.马赫学习。2001;45:5–32.
文章一个 Google Scholar一个
Statnikov A, Henaff M, Narendra V, Konganti K, Li Z, Yang L, et al.A comprehensive evaluation of multicategory classification methods for microbiomic data.微生物组。2013;1:11.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Deneke C, Rentzsch R, Renard BY.Paprbag: a machine learning approach for the detection of novel pathogens from NGS data.Sci Rep. 2017;7:39194.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Méric G, Mageiros L, Pensar J, Laabei M, Yahara K, Pascoe B, et al.Disease-associated genotypes of the commensal skin bacteriumStaphylococcus epidermidis。纳特社区。2018;9:5034.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Mageiros L, Méric G, Bayliss SC, Pensar J, Pascoe B, Mourkas E, et al.Genome evolution and the emergence of pathogenicity in avian大肠杆菌。纳特社区。2021;12:765.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Chen ML, Doddi A, Royer J, Freschi L, Schito M, Ezewudo M, et al.Beyond multidrug resistance: leveraging rare variants with machine and statistical learning models inMycobacterium tuberculosisresistance prediction.ebiomedicine。2019;43:356–69.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Walker H, et al.Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin binding protein (PBP) sequences.BMC基因组学。2017;18:621.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Arning N, Sheppard SK, Bayliss S, Clifton DA, Wilson DJ.Machine learning to predict the source of campylobacteriosis using whole genome data.PLOS基因。2021;17:e1009436 (Hughes D, editor.).
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Pascoe B, Futcher G, Pensar J, Bayliss SC, Mourkas E, Calland JK, et al.Machine learning to attribute the source ofCampylobacterinfections in the United States: a retrospective analysis of national surveillance data.J感染。2024;89:106265.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wheeler NE, Gardner PP, Barquist L. Machine learning identifies signatures of host adaptation in the bacterial pathogenSalmonella enterica。PLOS基因。2018;14:e1007333 (Didelot X, editor.).
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Zhang S, Li S, Gu W, Den Bakker H, Boxrud D, Taylor A, et al.Zoonotic Source Attribution ofSalmonella entericaSerotype Typhimurium Using Genomic Surveillance Data, United States.紧急感染疾病。2019;25.Available from:http://wwwnc.cdc.gov/eid/article/25/1/18-0835_article.htm. [cited 2025 Sept 2].
Beavan AJS, Domingo-Sananes MR, McInerney JO.Contingency, repeatability, and predictability in the evolution of a prokaryotic pangenome.美国科学学院。2024;121:e2304934120.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Mason L, Baxter J, Bartlett P, Frean M. Boosting Algorithms as Gradient Descent.Advances in Neural Information Processing Systems.MIT Press;1999. Available from:https://proceedings.neurips.cc/paper/1999/hash/96a93ba89a5b5c6c226e49b88973f46e-Abstract.html。Friedman JH.
Greedy function approximation: A gradient boosting machine.Ann Statist.2001;29.Available from:https://projecteuclid.org/journals/annals-of-statistics/volume-29/issue-5/Greedy-function-approximation-A-gradient-boosting-machine/10.1214/aos/1013203451.full. [cited 2025 Sept 2].
Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, et al.LightGBM: A Highly Efficient Gradient Boosting Decision Tree.Proceedings of the 31st International Conference on Neural Information Processing Systems.Red Hook, NY, USA: Curran Associates Inc;2017;3149–57 17.
Anahtar MN, Yang JH, Kanjilal S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: an Emerging Model for Translational Research.McAdam AJ, editor.J Clin Microbiol。2021;59:e01260–20.
Ramoneda J, Stallard-Olivera E, Hoffert M, Winfrey CC, Stadler M, Niño-GarcÃa JP, et al.Building a genome-based understanding of bacterial pH preferences.Sci Adv.2023;9:eadf8998.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Hopfield JJ.Neural networks and physical systems with emergent collective computational abilities.Proc Natl Acad Sci U S A. 1982;79:2554–8.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sheehan S, Song YS.Deep Learning for Population Genetic Inference.Chen K, editor.PLoS Comput Biol.2016;12:e1004845.
Li Y, Huang C, Ding L, Li Z, Pan Y, Gao X. Deep learning in bioinformatics: introduction, application, and perspective in the big data era.方法。2019;166:4–21.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Sejnowski TJ.The Deep Learning Revolution.The MIT Press;2018 Available from:https://direct.mit.edu/books/book/4111/The-Deep-Learning-Revolution. [cited 2025 Sept 2].
Lugo L, Hernández EB.A recurrent neural network approach for whole genome bacteria identification.Appl Artif Intell.2021;35:642–56.
文章一个 Google Scholar一个
Hasan MA, Lonardi S. Deeplyessential: a deep neural network for predicting essential genes in microbes.BMC生物信息学。2020;21:367.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Assaf R, Xia F, Stevens R. Detecting operons in bacterial genomes via visual representation learning.Sci Rep. 2021;11:2124.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Wiatrak M, Weimann A, Dinan A, Brbić M, Floto RA.Sequence-based modelling of bacterial genomes enables accurate antibiotic resistance prediction.Microbiology;2024 Available from:http://biorxiv.org/lookup/doi/10.1101/2024.01.03.574022. [cited 2025 Sept 2].
Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators.神经网。1989;2:359–66.
文章一个 Google Scholar一个
Zhang C, Bengio S, Hardt M, Recht B, Vinyals O. Understanding deep learning requires rethinking generalization.arxiv;2016. Available from:https://arxiv.org/abs/1611.03530。[cited 2025 Sept 2].
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al.注意就是您所需要的。Advances in Neural Information Processing Systems.2017;30.
Ouyang L, Wu J, Jiang X, Almeida D, Wainwright C, Mishkin P, et al.培训语言模型遵循人类反馈的说明。Adv Neural Inf Process Syst.2022;35:27730–44.
Holz HJ, Loew MH.Relative feature importance: A classifier-independent approach to feature selection.Machine Intelligence and Pattern Recognition.Elsevier;1994;473–87.Available from:https://linkinghub.elsevier.com/retrieve/pii/B9780444818928500468. [cited 2025 Sept 2].
Murdoch WJ, Singh C, Kumbier K, Abbasi-Asl R, Yu B. Definitions, methods, and applications in interpretable machine learning.美国科学学院。2019;116:22071–80.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
House of Commons Science, Innovation and Technology Committee.2023. The governance of artificial intelligence: interim report.Ninth Report of Session 2022–23.HC1769.https://committees.parliament.uk/publications/41130/documents/205611/default/
Nielsen EM, Fussing V, Engberg J, Nielsen NL, Neimann J. MostCampylobactersubtypes from sporadic infections can be found in retail poultry products and food animals.流行病感染。2006;134:758–67.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Garrett N, Devane ML, Hudson JA, Nicol C, Ball A, Klena JD, et al.Statistical comparison of弯曲杆菌空肠subtypes from human cases and environmental sources: comparison ofCampylobacter亚型。J Appl Microbiol.2007;103:2113–21.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, et al.Tracing the Source of Campylobacteriosis.Guttman DS, editor.PLOS基因。2008;4:e1000203.
Sheppard SK, Dallas JF, Strachan NJC, MacRae M, McCarthy ND, Wilson DJ, et al.Campylobactergenotyping to determine the source of human infection.临床感染。2009;48:1072–8.
文章一个 PubMed一个 Google Scholar一个
Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System.Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.San Francisco California USA: ACM;2016;785–94.Available from:https://dl.acm.org/doi/10.1145/2939672.2939785. [cited 2025 Sept 2].
Mackay TFC.The genetic architecture of quantitative traits.Annu Rev Genet.2001;35:303–39.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, et al.Virulent combinations of adhesin and toxin genes in natural populations of金黄色葡萄球菌。感染免疫。2002;70:4987–96.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Astle W, Balding DJ.Population Structure and Cryptic Relatedness in Genetic Association Studies.Statist Sci.2009;24.Available from: https://projecteuclid.org/journals/statistical-science/volume-24/issue-4/Population-Structure-and-Cryptic-Relatedness-in-Genetic-Association-Studies/10.1214/09-STS307.full. [cited 2025 Sept 2].
Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies.Nat Rev Genet。2010;11:459–63.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sheppard SK.Strain wars and the evolution of opportunistic pathogens.Curr Opin Microbiol.2022;67:102138.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Pearl J. Causal inference in statistics: An overview.Statist Surv.2009;3.Available from:https://projecteuclid.org/journals/statistics-surveys/volume-3/issue-none/Causal-inference-in-statistics-An-overview/10.1214/09-SS057.full. [cited 2025 Sept 2].
Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al.Causal associations between risk factors and common diseases inferred from GWAS summary data.纳特社区。2018;9:224.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, et al.Genome-wide association study identifies vitamin B5biosynthesis as a host specificity factor inCampylobacter。美国科学学院。2013;110:11923–7.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Earle SG, Wu C-H, Charlesworth J, Stoesser N, Gordon NC, Walker TM, et al.Identifying lineage effects when controlling for population structure improves power in bacterial association studies.NAT微生物。2016;1:16041.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. pyseer: a comprehensive tool for microbial pangenome-wide association studies.Stegle O, editor.生物信息学。2018;34:4310–2.
Young BC, Earle SG, Soeng S, Sar P, Kumar V, Hor S, et al.Panton-valentine leucocidin is the key determinant of金黄色葡萄球菌pyomyositis in a bacterial GWAS.Elife.2019;8:e42486.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Earle SG, Lobanovska M, Lavender H, Tang C, Exley RM, Ramos-Sevillano E, et al.Genome-wide association studies reveal the role of polymorphisms affecting factor H binding protein expression in host invasion by Neisseria meningitidis.Nassif X, editor.PLOS病原体。2021;17:e1009992.
Green AG, Yoon CH, Chen ML, Ektefaie Y, Fina M, Freschi L, et al.A convolutional neural network highlights mutations relevant to antimicrobial resistance inMycobacterium tuberculosis。纳特社区。2022;13:3817.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
The CRyPTIC Consortium.Genome-wide association studies of global Mycobacterium tuberculosis resistance to 13 antimicrobials in 10,228 genomes identify new resistance mechanisms.Ladner J, editor.Plos Biol。2022;20:e3001755.
Mosquera-Rendón J, Moreno-Herrera CX, Robledo J, Hurtado-Páez U. Genome-wide association studies (GWAS) approaches for the detection of genetic variants associated with antibiotic resistance: a systematic review.微生物。2023;11:2866.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW.Transforming clinical microbiology with bacterial genome sequencing.Nat Rev Genet。2012;13:601–12.
文章一个 CAS一个 PubMed一个 PubMed Central一个 Google Scholar一个
Walker TM, Cruz ALG, Peto TE, Smith EG, Esmail H, Crook DW.Tuberculosis is changing.Lancet Infect Dis.2017;17:359–61.
文章一个 PubMed一个 Google Scholar一个
Satta G, Lipman M, Smith GP, Arnold C, Kon OM, McHugh TD.Mycobacterium tuberculosisand whole-genome sequencing: how close are we to unleashing its full potential?Clin Microbiol Infect.2018;24:604–9.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE.Interpretation of Genetic Association Studies: Markers with Replicated Highly Significant Odds Ratios May Be Poor Classifiers.Abecasis GR, editor.PLOS基因。2009;5:e1000337.
Yang Y, Niehaus KE, Walker TM, Iqbal Z, Walker AS, Wilson DJ, et al.Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data.Birol I, editor.生物信息学。2018;34:1666–71.
Kouchaki S, Yang Y, Walker TM, Sarah Walker A, Wilson DJ, Peto TEA, et al.Application of machine learning techniques to tuberculosis drug resistance analysis.Wren J, editor.生物信息学。2019;35:2276–82.
Yang Y, Walker TM, Walker AS, Wilson DJ, Peto TEA, Crook DW, et al.DeepAMR for predicting co-occurrent resistance ofMycobacterium tuberculosis。Hancock J, editor.生物信息学。2019;35:3240–9.
Gröschel MI, Owens M, Freschi L, Vargas R, Marin MG, Phelan J, et al.Gentb: A user-friendly genome-based predictor for tuberculosis resistance powered by machine learning.基因组医学。2021;13:138.
文章一个 PubMed一个 PubMed Central一个 Google Scholar一个
The CRyPTIC Consortium and the 100,000 Genomes Project.Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing.N Engl J Med。2018;379:1403–15.
He G, Zheng Q, Shi J, Wu L, Huang B, Yang Y. Evaluation of WHO catalog of mutations and five WGS analysis tools for drug resistance prediction ofMycobacterium tuberculosisisolates from China.Georghiou SB, editor.微生物谱。2024;12:e03341–23.
Ferrari E, Retico A, Bacciu D. Measuring the effects of confounders in medical supervised classification problems: the confounding index (CI).Artif Intell Med。2020;103:101804.
文章一个 PubMed一个 Google Scholar一个
Ribeiro MT, Singh S, Guestrin C. “Why Should I Trust You?â€: Explaining the Predictions of Any Classifier.Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.San Francisco California USA: ACM;2016;1135–44.Available from:https://dl.acm.org/doi/10.1145/2939672.2939778. [cited 2025 Sept 2].
Lundberg S, Lee S-I.A Unified Approach to Interpreting Model Predictions.arxiv;2017 Available from:https://arxiv.org/abs/1705.07874. [cited 2025 Sept 2].
Meyes R, Lu M, Waubert de Puiseau C, Meisen T. Ablation studies to uncover structure of learned representations in artificial neural networks.Proceedings of the International Conference on Artificial Intelligence (ICAI).Athens, Greece: CSREA Press;2019 Available from:https://www.researchgate.net/publication/334871296_Ablation_Studies_to_Uncover_Structure_of_Learned_Representations_in_Artificial_Neural_Networks. [cited 2025 Sept 2].
Callaway E. How generative AI is building better antibodies.自然。2023;d41586–023–01516-w.
118.Callaway E. ‘ChatGPT for CRISPR’ creates new gene-editing tools.自然。2024;629:272–272.
文章一个 CAS一个 PubMed一个 Google Scholar一个
Tang X, Dai H, Knight E, Wu F, Li Y, Li T, et al.A survey of generative AI for de novo drug design: new frontiers in molecule and protein generation.Briefings in Bioinformatics.2024;25:bbae338
Winnifrith A, Outeiral C, Hie BL.Generative artificial intelligence for de novo protein design.Current Opinion in Structural Biology.2024;86:102794
致谢
不适用。
同行评审信息
Claudia Feng was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.The peer-review history is available in the online version of this article.
资金
SKS was supported by an Ineos Oxford Institute grant;Wellcome Trust grant 088786/C/09/Z, and UKRI grants MR/L015080/1, MR/V001213/1, MR/S009264/1, and MR/T030062/1.NA was supported by a BBSRC scholarship BB/M011224/1.DWE was supported by the NIHR Oxford Biomedical Research Centre, the NIHR Health Protection Research Unit in Healthcare Associated Infection and Antimicrobial Resistance and by a Robertson Fellowship.DJW was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society 101237/Z/13/B and by a Robertson Fellowship.表达的观点是作者的观点,不一定是NIHR或卫生和社会护理部的观点。
Ineos Oxford Institute, Wellcome (088786/C/09/Z, 101237/Z/13/B), UK Research and Innovation (MR/L015080/1), Biotechnology and Biological Sciences Research Council (BB/M011224/1), NIHR Oxford Biomedical Research Centre, NIHR Health Protection Research Unit in Healthcare Associated Infection and Antimicrobial Resistance, Robertson Foundation, Royal Society (101237/Z/13/B).
道德批准并同意参加
不适用。
同意出版不适用。
竞争利益
作者没有宣称没有竞争利益。
附加信息
Publisher’s Note
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。
权利和权限
开放访问
本文允许以任何媒介或格式的使用,共享,适应,分发和复制的方式获得创意共享归因4.0国际许可证,只要您对原始作者和来源提供适当的信誉,就可以提供与创意共享许可证的链接,并指出是否进行了更改。
The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material.如果文章的创意共享许可中未包含材料,并且您的预期用途不得由法定法规允许或超过允许的用途,则需要直接从版权所有者那里获得许可。要查看此许可证的副本,请访问http://creativecommons.org/licenses/by/4.0/。重印和权限
引用本文
Sheppard, S.K., Arning, N., Eyre, D.W.
等。Machine learning and statistical inference in microbial population genomics.基因组生物26 , 313 (2025).https://doi.org/10.1186/s13059-025-03775-4
已收到:
公认:
出版:
doi:https://doi.org/10.1186/s13059-025-03775-4