Abstract
Purpose
Predicting clinical complete response (CR) to neoadjuvant chemotherapy (NAC) in patients with muscle-invasive bladder cancer (MIBC) remains a clinical challenge. Existing CT-based radiomics studies have shown promise, but MRI-derived radiomics using machine learning (ML) has not been systematically explored. This study aimed to develop and validate ML-based radiomics models using multiparametric MRI and clinical data to predict CR in MIBC patients receiving NAC.
Materials and methods
MIBC patients eligible for platinum-based NAC were prospectively included. Tumor regions were manually segmented from pre-treatment MRI sequences (CE-T1WI, T2WI, DWI, ADC maps). Radiomics features and clinical variables were extracted. Least Absolute Shrinkage and Selection Operator (LASSO) was used for feature selection, and multiple ML classifiers were trained using stratified fivefold cross-validation. The area under the receiver operating characteristic curve (AUC-ROC), sensitivity, specificity, precision, and F1 scores were calculated.
Results
Among 52 patients, 19 (36.5%) achieved CR. Of 177 extracted features, CE-T1WI-derived models achieved the best performance. The support vector machine (SVM) yielded the highest AUC-ROC of 0.88, with sensitivity, specificity, and precision of 0.82, 0.79, and 0.79, respectively. The K-Nearest Neighbors (KNN) model performed comparably (AUC = 0.87). Clinical feature-based models also performed strongly (RF, AUC = 0.86).
Conclusions
ML-based radiomics models derived from multiparametric MRI sequences and clinical features hold substantial potential for predicting clinical CR to the NAC in MIBC patients. These results suggest that MR images can provide reliable insights into treatment response, offering a noninvasive and effective tool for clinical decision-making. This is the first prospective ML-based MRI radiomics study in this domain. We present this work as a proof-of-concept requiring external multicenter validation in a larger dataset to confirm these findings.
Introduction
Muscle-invasive bladder cancer (MIBC) is one of the most common malignant tumors of the genitourinary system, with a considerable recurrence rate [1]. Neoadjuvant chemotherapy (NAC) can reduce the burden of micro-metastasis and improve the prognosis for patients who are sensitive to platinum-based chemotherapy [2]. Nevertheless, NAC for non-responders may result in unnecessary delays in surgical treatment and chemotherapy-related toxicity [3]. Therefore, a tool for the early assessment of tumor response to NAC is crucial for the management of patients with bladder cancer.
Artificial intelligence (AI) algorithms can greatly improve the diagnosis, treatment, and prediction of outcomes in healthcare. [4]. By employing machine learning (ML) algorithms, we can uncover patterns in medical data that were previously difficult to identify, thereby improving the management of different tumor types, including urological cancers. [5]. In recent years, researchers have shown the use of ML-based radiomics for the prediction of tumor response to the NAC in different types of cancer [6,7,8,9]. The key concept behind radiomics is that the image contains more information than visual perception. This hidden information can be extracted using complex algorithms to probe tumor heterogeneity [10].
Some novel approaches have been employed in various cancers to enhance the performance and optimize model classification [11,12,13,14,15,16]. Furthermore, the performance of radiomics models can vary significantly depending on the imaging modality used [7]. Previous AI-based research in the area of response prediction for MIBC patients has relied on radiomics features extracted from CT images [17,18,19]. Although CT scans provide high-resolution images, MRI is particularly effective at providing detailed images of soft tissues. To our knowledge, no MRI-based radiomics studies in patients with MIBC utilize ML algorithms to predict tumor response to NAC. Considering this research gap, this study develops the first prospective ML-based MRI radiomics models for predicting clinical CR to NAC in MIBC, integrating both radiomics and clinical features. Our pipeline differs from prior works by prospectively enrolling patients, incorporating multiple MRI sequences, providing a Rad-Score formula for transparent interpretation, and comparative analysis across multiple ML classifiers.
Methods
Study design and protocol
This prospective cohort study was conducted at two tertiary hospitals between July 2019 and July 2021. It involved consecutive male and female patients diagnosed with MIBC at clinical stages T2 to T4a, who had previously undergone transurethral resection of bladder tumor (TURBT). All participants were eligible for platinum-based NAC and had a performance status ranging from 0 to 2. The inclusion criteria also required patients to have sufficient baseline bone marrow and liver function. Gemcitabine was administered to patients in combination with either cisplatin or carboplatin in two chemotherapy regimens. The treatment protocol has been previously published [20] and is detailed in Supplementary Methods (Supplementary File 1). We reported this study based on the TRIPOD statement. All patients provided written informed consent. As a rule of thumb, at least 10 samples per feature were considered in the final radiomics models [21]. Figure 1 illustrates the comprehensive workflow of the study, outlining each stage of the research process.
Overall workflow of the study. Block diagram summarizing the study design and analytic pipeline. Steps include patient selection (prospective cohort of MIBC treated with NAC), multiparametric MRI acquisition (CE-T1WI, T2WI, DWI, ADC), tumor segmentation, radiomics feature extraction, feature selection with LASSO, model training with multiple machine learning classifiers under stratified fivefold cross-validation, and performance evaluation. This schematic emphasizes the transparency and reproducibility of the modeling process
Outcome and assessment
The main objective of the study was to assess the clinical CR to chemotherapy [22]. A patient was deemed to have achieved a clinical CR if cystoscopy, along with tumor site biopsy, urine cytology, and imaging, showed no evidence of a primary tumor (T0). This evaluation took place four weeks after treatment completion through cystoscopy and imaging procedures. The cystoscopy was conducted by an independent urologist unaware of the treatment details. Only patients undergoing pre-treatment MRI and post-treatment cystoscopy were included in the available-case analysis.
MR Image acquisition and ROI delineation
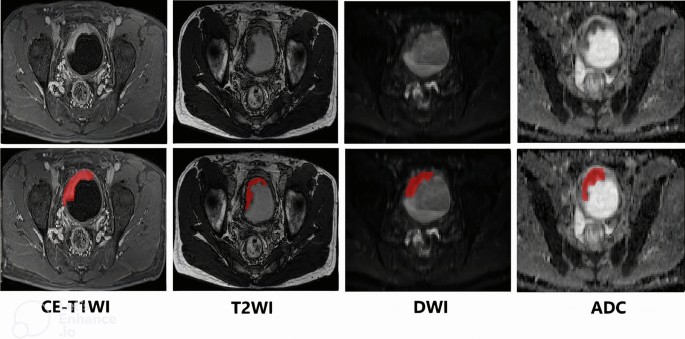
A detailed protocol for MR image acquisition was published earlier [20] and is summarized in Supplementary Methods (Supplementary File 1). A radiologist with over 10 years of experience reading MRIs who was blinded to the histopathologic results and clinical outcomes of the patients manually drew the regions of interest (ROI) on each tumor in various pre-treatment MR images including contrast-enhanced T1-weighted imaging (CE-T1WI), T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), and apparent diffusion coefficient maps (ADC maps), starting from the first image showing tumor presence and continuing through to the last image. Figure 2 illustrates a representative example of tumor segmentation on imaging, highlighting the delineation of the tumor region for analysis. The entire segmentation process was performed using ITK-SNAP software v.4.2.0 [23].
Radiomics feature extraction
In this study, we used a Python package for quantitative analysis of medical images, Medical Image Radiomics Processor (MIRP) [24] for radiomics feature extraction. The MIRP is designed to extract standardized radiomics and offers a consistent framework for feature extraction, ensuring reproducibility and reliability in radiomic studies based on the Image Biomarker Standardization Initiative (IBSI) protocols. The feature sets are defined in Supplementary File 1.
Feature selection and building Rad-Score
In this study, we applied the Least Absolute Shrinkage and Selection Operator (LASSO) for feature selection to identify the most predictive radiomics features while reducing dimensionality and redundancy. We developed radiomics signatures using LASSO by selecting the five most important features based on their nonzero coefficients. These features were combined into a radiomics score (Rad-Score). More details regarding the feature selection and building Rad-Score are summarized in Supplementary File 1.
Model development and validation
Several ML algorithms were utilized to develop predictive models, including K-nearest neighbors (KNN), logistic regression (LR), random forest (RF), support vector machine (SVM), LightGBM (LGBM), and XGBoost (XGB). Each algorithm was tuned for optimal performance. For hyperparameter tuning, GridSearchCV was utilized to perform an exhaustive search over predefined grids of hyperparameters for each ML model. The list of hyperparameters and their ranges is summarized in Supplementary Table 11 (Supplementary File 2). Also, parallelization was enabled by setting n_jobs = −1, allowing the process to leverage all available CPU cores and significantly reduce computation time.
In this study, a stratified K-fold cross-validation approach with 5 splits (~ 41 patients for training and ~ 11 patients for validation) was employed, maintaining balanced class proportions across training and validation folds.
Model evaluation and performance
The model performance evaluation was based on a comprehensive set of metrics to ensure a robust analysis of classification effectiveness. The primary scoring metric was the area under the receiver operating characteristic curve (AUC-ROC), selected for its ability to balance sensitivity and specificity. For ROC/AUC computation, fivefold cross-validation was employed. We aggregated out-of-fold predicted probabilities across all fivefold to produce a single consolidated ROC curve per model/feature set. This avoids optimistic bias that arises from plotting the ROC on training data. Additional metrics used in our study are summarized in Supplementary File 1.
Limitations of the experimental setup
This study was based on a modest prospective cohort from two centers, and external validation was not performed. Manual segmentation may also have introduced variability. The use of a single expert reader and the limited generalizability should also be considered. These limitations are further discussed in the Discussion section.
Results
Patient cohort
Of the 62 patients recruited, 52 with available pre-treatment MRI and response status were included in our available-case analyses. Among these, 19 patients (36.5%) achieved CR, while 33 (63.5%) did not. The characteristics of patients are indicated in Table 1. The majority of patients were male, accounting for 84.2% of the CR group and 87.9% of the non-CR group. In terms of tumor characteristics, most patients had high-grade tumors, comprising 84.2% in the CR group and 87.9% in the group without CR. The mean baseline tumor volume was smaller for patients with CR (6.3cm3) than those without CR (22.9cm3). Additionally, patients with CR were more likely to have negative nodal status (63.2%) than those without CR (36.4%).
Extracted features
Radiomic features were extracted from various image series, including CE-T1WI, T2WI, DWI, and ADC map, to assess their predictive value for clinical CR to the NAC. These features included a range of first-order statistics, texture-based metrics, and shape descriptors, capturing diverse tumor characteristics such as intensity distribution, spatial heterogeneity, and geometric complexity. The 177 radiomics features extracted are listed in Supplementary File 3. In addition to imaging-derived features, clinical variables including age, sex, tumor volume, T stage, grade, nodal status, and chemotherapy regimen were considered clinical features. In the feature selection process, the LASSO algorithm was employed to identify the five most predictive features for each model.
To quantify the predictive value of radiomics and clinical features, a Rad-Score was calculated using a linear combination of selected features. The Rad-Score formula for CE-T1WI, which was identified as the best image sequence due to its higher AUC-ROC, is as follows:
$$\begin{aligned} {\text{Rad}} - {\text{Score}}_{{{\text{CE}} - {\text{T1WI}}}} & = 0.{3654} + \left( {{2}.{\text{6636e}} - 0{1} \times {\text{ih}}\_{\text{p1}}0\_{\text{fbn}}\_{\text{n16}}} \right) + \left( {{2}.{\text{5514e}} - 0{1} \times {\text{morph}}\_{\text{vol}}\_{\text{dens}}\_{\text{aabb}}} \right) \\ & \quad + \left( {{2}.0{\text{787e}} - 0{1} \times {\text{morph}}\_{\text{vol}}\_{\text{dens}}\_{\text{aee}}} \right) + \left( {{1}.{\text{6927e}} - 0{1} \times {\text{cm}}\_{\text{clust}}\_{\text{shade}}\_{\text{d1}}\_{\text{2d}}\_{\text{s}}\_{\text{mrg}}\_{\text{fbn}}\_{\text{n16}}} \right) \\ & \quad + ({1}.{\text{5351e}} - 0{1} \times {\text{ivh}}\_{\text{v25}}) \\ \end{aligned}$$
This equation integrates key imaging features weighted by their coefficients, emphasizing their relative contribution to CR prediction. The Rad-Score equations for different models and the Rad-Score calculated for each patient are indicated in Supplementary File 4. The Rad-Score formulation facilitates failure-case analysis by providing transparent feature contributions for each case. Failure cases often involved heterogeneous tumors or borderline feature values. In misclassified patients, we observed atypical combinations of feature values that deviated from the expected patterns in responders and non-responders. For example, false-negative cases frequently showed Rad-Scores driven by low-volume morphology features, despite clinical CR being achieved. Conversely, several false-positive cases exhibited high heterogeneity indices and elevated vascularity features, mimicking the profile of true responders.
Performance of models
Figure 3 indicates the ROC curves as the main index for evaluating the predictive performance of various ML classifiers applied to CE-T1WI, T2WI, DWI, ADC maps, and clinical models. AUC-ROC values greater than 0.80 are typically regarded as clinically useful, whereas values below 0.80 are considered to have limited clinical value [25]. Supplementary File 5 summarizes 95% confidence intervals (95%CIs) and P values for all AUC-ROCs in our study.
ROC diagrams of different sets of features: a CE-T1WI, b T2WI, c DWI, d ADC map, and e clinical. ROC curves generated from aggregated out-of-fold predictions under stratified fivefold cross-validation. This figure demonstrates the discriminative ability of the models for predicting NAC response. AUC-ROC values greater than 0.80 are typically regarded as clinically useful, whereas values below 0.80 are considered to have limited clinical value
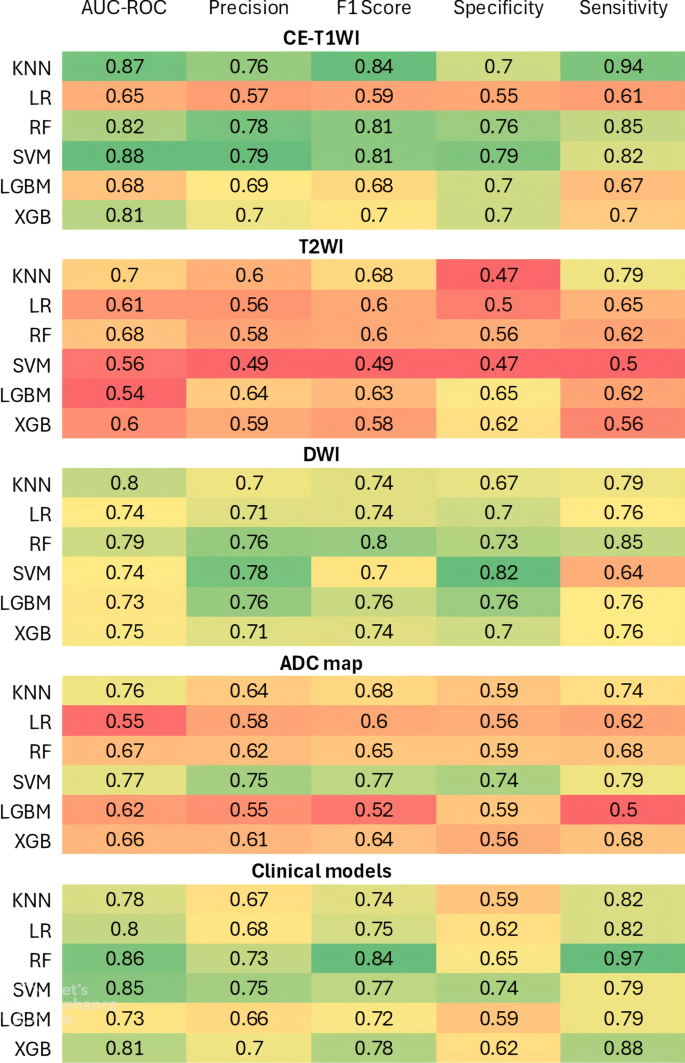
Upon comparing different image series, CE-T1WI demonstrated the strongest predictive capacity, particularly with the SVM classifier, which achieved an AUC-ROC of 0.88 (95%CI: 0.75–0.95; P < 0.001). A model based on the KNN classifier performs second best when using the CE-T1WI sequence, achieving an AUC-ROC of 0.87 (95%CI: 0.77–0.95; P < 0.001). The LR model showed the lowest performance for CE-T1WI, with an AUC-ROC of 0.65 (95%CI: 0.51–0.79; P = 0.022), indicating a poor predictive value.
The T2WI-derived models showed lower predictive performance across most classifiers compared to models derived from other MRI image series. The KNN classifier achieved the highest AUC-ROC of 0.70 (95%CI: 0.57–0.82; P = 0.002) for this sequence. Other classifiers yielded poor results for T2WI, with AUC-ROC values ranging from 0.54 to 0.68. Overall, T2WI was found to be a weak sequence for predicting response.
For the DWI sequence, all classifiers demonstrated moderate predictive performance. KNN and RF emerged as top performers, achieving AUC-ROC values of 0.80 (95%CI: 0.69–0.90; P < 0.001) and 0.79 (95%CI: 0.66–0.90; P < 0.001), respectively. Other algorithms provided comparable results, with AUC-ROC values ranging from 0.73 to 0.75.
ADC maps showed poor to moderate performance. The SVM achieved the highest AUC-ROC of 0.77 (95%CI: 0.63–0.87; P < 0.001) among classifiers, similar to those obtained for the DWI image series. The KNN algorithm followed, with an AUC-ROC value of 0.76 (95%CI: 0.63–0.87; P < 0.001). Other classifiers yielded poor performance, achieving AUC-ROCs between 0.55 and 0.67.
Interestingly, clinical features yielded moderate to good performance, demonstrating their standalone predictive value. The RF model achieved the highest AUC-ROC of 0.86 (95%CI: 0.77–0.94; P < 0.001) among all classifiers. The SVM also performed strongly, with an AUC-ROC of 0.85 (95%CI: 0.72–0.93; P < 0.001). The KNN, LR, and XGB showed slightly lower AUC-ROC values of 0.78 (95%CI: 0.66–0.88; P < 0.001), 0.80 (95%CI: 0.68–0.89; P < 0.001), and 0.81 (95%CI: 0.70–0.91; P < 0.001), respectively. The LGBM model showed the lowest performance for clinical features, with an AUC-ROC of 0.73 (95%CI: 0.60–0.84, P < 0.001).
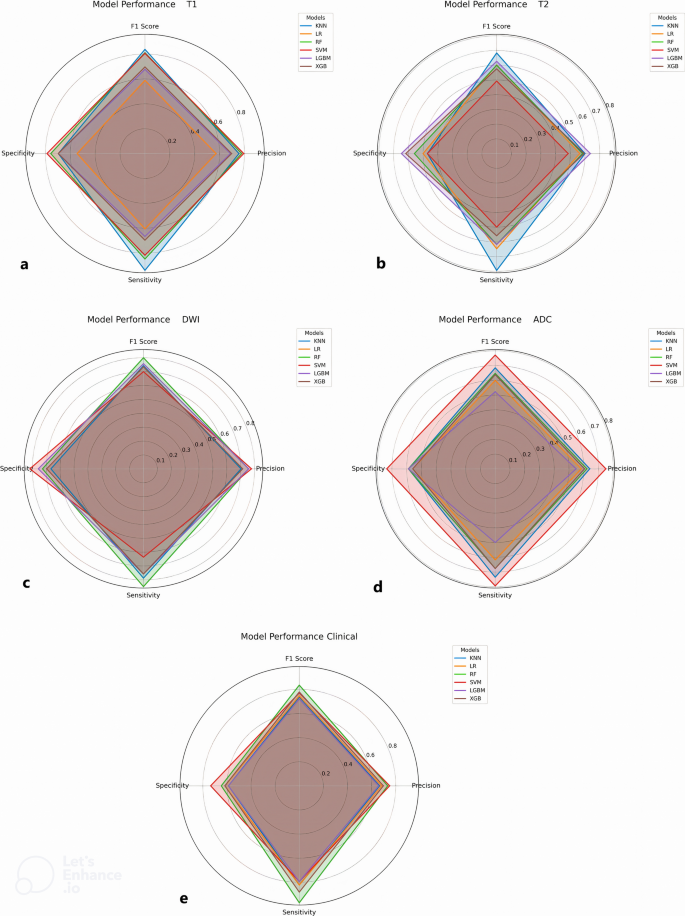
Figure 4 indicates the performance of different models with radar plots summarizing key performance metrics, including precision, sensitivity, specificity, and F1 score. These plots highlight the robustness of the classifiers when applied to CE-T1WI-derived radiomics and clinical features while demonstrating variability in performance across different models.
Radar plots of different sets of features: a CE-T1WI, b T2WI, c DWI, d ADC map, and e clinical. The plots show sensitivity, specificity, precision, and F1 score for each machine learning classifier across different image series and clinical features. These metrics account for class imbalance and provide a fair assessment of model performance beyond AUC
Moreover, Fig. 5 provides a comparative visualization of the predictive performance of ML classifiers across different image series and clinical features using a heatmap plot. The metrics assessed include AUC-ROC, precision, F1 score, specificity, and sensitivity, providing a comprehensive evaluation of classifier accuracy, reliability, and robustness across different feature sets. In summary, classification was performed at the patient level, with one response label assigned per patient. All cross-validation folds were stratified to preserve the proportion of responders and non-responders. For classifiers that support imbalance handling, we applied class weighting during training to reduce bias toward the majority class. In addition to the primary AUC-ROC metric, we report balanced performance measures, including sensitivity, specificity, precision, and F1 score, which are summarized in Supplementary File 6.
Heatmap depicting the predictive performance of machine learning classifiers for various MR image series and clinical features. Comparison of classifier performance when trained on different feature sets. CE-T1WI produced the highest discriminative performance, highlighting the importance of contrast-enhanced imaging for response prediction
The results highlight notable variations in model performance depending on the input data type and the algorithm employed, underscoring the importance of modality-specific optimization. Regarding the performance of the ML algorithms used, KNN, SVM, and RF achieved the best results across most models, while LR and LGBM showed poor performance. A more complete set of metrics for assessing model performance is outlined in Supplementary File 6.
Discussion
Bladder cancer is a significant public health concern due to its high recurrence rate and considerable variability in patient outcomes. Accurately predicting how patients will respond to chemotherapy is critical, as it can guide clinicians in personalizing treatment plans and avoiding unnecessary toxicities [3]. Thus, there is an increasing demand for noninvasive methods to predict treatment outcomes, which could improve the quality of life for bladder cancer patients and optimize therapeutic decision-making.
ML methods, combined with MRI radiomics, have emerged as promising tools in oncology for noninvasive prediction of treatment response [5]. Radiomics enables the extraction of quantitative features from medical imaging modalities that capture tumor heterogeneity and other robust features not visible to the human eye [10]. By applying advanced ML algorithms, researchers aim to leverage these features to create models that can predict treatment efficacy before chemotherapy [26]. These models can benefit bladder cancer patients, providing a pathway toward individualized treatment and potentially better survival rates [5]. Novel contributions in our study include the incorporation of the multiparametric MRI sequences, the application of Rad-Score feature engineering, and comparative analysis across multiple ML classifiers.
In a previous study on MIBC patients, Cha et al. [19] developed CT scan-based radiomics models to predict NAC response using 91 radiomics features. Testing the best model achieved an AUC value of 0.77 in predicting CR after treatment. In a subsequent study by the same group, Wu et al. [17] utilized transfer learning to train a model using CT scans from 123 MIBC patients undergoing NAC, resulting in a test AUC of 0.79. In another study by Choi et al. [18], a CT scan-derived radiomics signature was developed to predict pathological CR with an RF classifier, achieving a good performance in the training dataset (AUC, 0.85) and moderate performance in the validation (AUC, 0.75) set. A comparative analysis of these prior radiomics studies and the present work is provided in Supplementary Table 2 (Supplementary File 2), summarizing modality, study design, methods, best reported AUC, and limitations. This table situates our MRI-derived AUC within the context of earlier CT-based studies, highlighting its improved discriminatory performance and the added value of MRI sequences for soft tissue characterization. Furthermore, Supplementary Table 3 (Supplementary File 2) presents an architectural comparison between our Rad-Score plus classical ML pipeline and commonly used attention-based pipelines, considering input type, feature extraction, interpretability, data requirements, risk of overfitting, and computational cost. This table illustrates that the methodological novelty of our study lies in the prospective acquisition of MRI data, standardized IBSI-compliant feature extraction (MIRP), transparent Rad-Score equations for clinical interpretability, and a systematic multi-classifier evaluation with robust cross-validation.
In this study, we developed ML-based radiomic models using four MR image series (CE-T1WI, T2WI, DWI, and ADC map) alongside clinical features to predict clinical CR to the NAC in patients with MIBC. The results demonstrated that different image series and clinical features offer varying degrees of predictive power.
Among various image series, CE-T1WI exhibited superior predictive performance, with classifiers such as SVM and KNN achieving the highest AUC-ROC values (0.88 and 0.87, respectively). This suggests that this sequence captures key tumor characteristics, such as tumor shape, margins, extension, heterogeneity, and vascular permeability, which are critical for assessing treatment response. In contrast, T2WI showed poor predictive power, indicating that while this sequence contributes valuable information, it may require additional optimization or feature engineering to enhance its utility. Also, the varying performance metrics across different sequences underscore the necessity of multimodal approaches, as demonstrated in large studies that integrated various imaging techniques to enhance predictive power [8].
Using clinical features for response prediction resulted in good model performance, with RF and SVM classifiers achieving the highest predictive accuracy. Baseline clinical characteristics including tumor volume, T stage, and nodal status remain fundamental contributors to predictive models. These findings align with previous radiomics studies that highlight the inclusion of clinical data in predictive models [27, 28]. Regarding statistical considerations, integrating clinical and radiomics features into a single model necessitates a relatively large dataset. Given the limited size of our primary dataset, we developed distinct models for different clinical and imaging features in this study.
Among the different classifiers used in our study, the SVM algorithm consistently outperformed others, achieving an AUC of 0.88. This aligns with existing studies that support the utility of SVM in radiomics, emphasizing its capability to capture complex patterns in imaging data [29,30,31,32]. For instance, a study by Du et al. [33] demonstrated that SVM, when applied to radiomic features extracted from pre-treatment MRI, could significantly predict the treatment response of brain metastasis patients to stereotactic radiosurgery, further highlighting the robustness of this classifier in oncology. Additionally, Ji et al. [34] found that SVM classifiers effectively predict lymph node metastasis in patients with bladder urothelial carcinoma treated with radical cystectomy, reinforcing the findings of this analysis. Similar to our findings, Cai et al. [8] emphasized the promising role of MRI-based radiomics using SVM classifiers in predicting the response to NAC of patients with cervical cancer. Their results demonstrated that MRI-derived radiomics could identify key features related to tumor heterogeneity and vascular permeability, achieving an AUC of 0.86 for treatment response prediction. Similarly, our study’s AUC of 0.88 for CE-T1WI-based radiomics with SVM corroborates the high discriminatory power of MRI sequences for predicting CR.
In our study, the LGBM classifier demonstrated the lowest predictive performance for assessing chemotherapy response across T2WI, DWI, and clinical features. Several factors may account for this result. LGBM is known for its efficiency with large datasets due to its gradient-based one-sided sampling and exclusive feature bundling, which makes it highly effective in cases with abundant, high-dimensional data. However, for smaller or imbalanced datasets, such as those often encountered in radiomics studies, the LGBM algorithm may not capture subtle patterns effectively, potentially leading to underperformance. Comparing this outcome with previous research, some studies have reported the favorable performance of LGBM for predicting early complications following radical gastrectomy and breast cancer classification, particularly when extensive datasets are available [35, 36]. This contrast suggests that LGBM’s applicability may be context-dependent, and alternative models like SVM or RF might be more suitable for small-scale, high-dimensional radiomics data. Future research should consider further investigation into dataset size and balance requirements for LGBM’s optimal use in predictive modeling.
Clinical implication
Our findings suggest that our MRI-based radiomics model, combined with the Rad-Score and ML classifiers, may serve as a valuable decision-support tool in the management of MIBC patients. By enabling noninvasive, pre-treatment prediction of response to NAC at the patient level, this approach could help clinicians identify candidates who are more likely to benefit from NAC and, conversely, spare non-responders from unnecessary toxicity and delays in definitive therapy. The transparency of the Rad-Score further facilitates interpretability, allowing physicians to understand which imaging features contribute to an individual prediction. While additional validation in larger, multicenter cohorts is required, the model has the potential to support personalized treatment planning and to complement multidisciplinary decision-making.
Limitations
Despite these promising results, this study has several limitations. First, the sample size, although sufficient for initial model development, may limit the generalizability of the findings to larger populations. Second, external validation on independent cohorts is necessary to ensure the reproducibility and clinical applicability of the developed models. Third, while class weighting, stratified cross-validation, and LASSO feature selection helped mitigate overfitting and imbalance, these techniques may not fully eliminate bias in small datasets. Fourth, consideration should be given to the relatively wide range of the 95% CIs in some models. A narrow CI suggests that the AUC value is probably accurate, whereas a wide CI indicates that the AUC value is less reliable [25].
Future directions
Future studies may benefit from the integration of advanced deep learning methods and the use of longitudinal imaging to capture temporal dynamics throughout NAC. Furthermore, establishing multi-institutional collaborations will enable access to larger datasets and support external validation, thereby strengthening the robustness and clinical applicability of ML-based radiomics models.
Conclusions
This study showed the potential of ML-based radiomics models, derived from multiparametric MRI sequences and clinical features, for predicting clinical CR to NAC in MIBC patients. The CE-T1WI-derived models were developed as the most influential predictors, with classifiers such as SVM and KNN consistently demonstrating high predictive performance. To our knowledge, this is the first study to establish a strong basis for applying ML-based radiomics as a noninvasive approach to guide treatment decisions and enhance patient outcomes in bladder cancer management. While promising, external multicenter validation is necessary to confirm these findings.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Mari A, Campi R, Tellini R, Gandaglia G, Albisinni S, Abufaraj M, Hatzichristodoulou G, Montorsi F, van Velthoven R, Carini M et al (2018) Patterns and predictors of recurrence after open radical cystectomy for bladder cancer: a comprehensive review of the literature. World J Urol 36(2):157–170
Hamid A, Ridwan FR, Parikesit D, Widia F, Mochtar CA, Umbas R (2020) Meta-analysis of neoadjuvant chemotherapy compared to radical cystectomy alone in improving overall survival of muscle-invasive bladder cancer patients. BMC Urol 20(1):158
Schardt J, Roth B, Seiler R (2019) Forty years of cisplatin-based chemotherapy in muscle-invasive bladder cancer: are we understanding how, who and when? World J Urol 37(9):1759–1765
Maleki Varnosfaderani S, Forouzanfar M (2024) The role of AI in hospitals and clinics: transforming healthcare in the 21st century. Bioengineering. https://doi.org/10.3390/bioengineering11040337
Pak S, Park SG, Park J, Cho ST, Lee YG, Ahn H (2024) Applications of artificial intelligence in urologic oncology. Investig Clin Urol 65(3):202–216
McAnena P, Moloney BM, Browne R, O’Halloran N, Walsh L, Walsh S, Sheppard D, Sweeney KJ, Kerin MJ, Lowery AJ (2022) A radiomic model to classify response to neoadjuvant chemotherapy in breast cancer. BMC Med Imaging 22(1):225
Zhang L, Wang Y, Peng Z, Weng Y, Fang Z, Xiao F, Zhang C, Fan Z, Huang K, Zhu Y et al (2022) The progress of multimodal imaging combination and subregion based radiomics research of cancers. Int J Biol Sci 18(8):3458–3469
Cai Z, Li S, Xiong Z, Lin J, Sun Y (2024) Multimodal MRI-based deep-radiomics model predicts response in cervical cancer treated with neoadjuvant chemoradiotherapy. Sci Rep 14(1):19090
Chen BY, Xie H, Li Y, Jiang XH, Xiong L, Tang XF, Lin XF, Li L, Cai PQ (2022) MRI-based radiomics features to predict treatment response to neoadjuvant chemotherapy in locally advanced rectal cancer: a single center, prospective study. Front Oncol 12:801743
Parekh V, Jacobs MA (2016) Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev 1(2):207–226
Hekmat A, Zuping Z, Bilal O, Khan SUR (2025) Differential evolution-driven optimized ensemble network for brain tumor detection. Int J Mach Learn Cybern 16(9):6447–6472
Rehman Khan SU, Asif S, Zhao M, Zou W, Li Y, Xiao C (2026) ShallowMRI: a novel lightweight CNN with novel attention mechanism for multi brain tumor classification in MRI images. Biomed Signal Process Control 111:108425
Bilal O, Hekmat A, Khan SUR (2025) Automated cervical cancer cell diagnosis via grid search-optimized multi-CNN ensemble networks. Netw Model Anal Health Inform Bioinform 14(1):67
Khan SUR, Asim MN, Vollmer S, Dengel A (2025) AI-Driven Diabetic Retinopathy Diagnosis Enhancement through Image Processing and Salp Swarm Algorithm-Optimized Ensemble Network. arXiv preprint arXiv:250314209
Khan SUR, Asim MN, Vollmer S, Dengel A (2025) FOLC-Net: A Federated-Optimized Lightweight Architecture for Enhanced MRI Disease Diagnosis across Axial, Coronal, and Sagittal Views. arXiv preprint arXiv:250706763
Maqsood H, Khan SUR (2025) MeD-3D: A Multimodal Deep Learning Framework for Precise Recurrence Prediction in Clear Cell Renal Cell Carcinoma (ccRCC). arXiv preprint arXiv:250707839
Wu E, Hadjiiski LM, Samala RK, Chan HP, Cha KH, Richter C, Cohan RH, Caoili EM, Paramagul C, Alva A et al (2019) Deep learning approach for assessment of bladder cancer treatment response. Tomography 5(1):201–208
Choi SJ, Park KJ, Heo C, Park BW, Kim M, Kim JK (2021) Radiomics-based model for predicting pathological complete response to neoadjuvant chemotherapy in muscle-invasive bladder cancer. Clin Radiol 76(8):627.e613-627.e621
Cha KH, Hadjiiski L, Chan H-P, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C, Samala RK (2017) Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep 7(1):8738
Razzaghdoust A, Jafari A, Mahdavi A, Mofid B, Basiri A (2025) Diffusion-weighted MRI-derived ADC and tumor volume as predictive imaging markers for neoadjuvant chemotherapy response in muscle-invasive bladder cancer. BMC Med Imaging 25(1):3
Shur JD, Doran SJ, Kumar S, Ap Dafydd D, Downey K, O’Connor JPB, Papanikolaou N, Messiou C, Koh DM, Orton MR (2021) Radiomics in oncology: a practical guide. Radiographics 41(6):1717–1732
Wu J, Xie R-Y, Cao C-Z, Shang B-Q, Shi H-Z, Shou J-Z (2022) Disease management of clinical complete responders to neoadjuvant chemotherapy of muscle-invasive bladder cancer: a review of literature. Front Oncol. https://doi.org/10.3389/fonc.2022.816444
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3d active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Zwanenburg A, Löck S (2024) MIRP: a Python package for standardised radiomics. J Open Source Softw 9(99):6413
Çorbacıoğlu ŞK, Aksel G (2023) Receiver operating characteristic curve analysis in diagnostic accuracy studies: a guide to interpreting the area under the curve value. Turk J Emerg Med 23(4):195–198
Feretzakis G, Juliebø-Jones P, Tsaturyan A, Sener TE, Verykios VS, Karapiperis D, Bellos T, Katsimperis S, Angelopoulos P, Varkarakis I (2024) Emerging trends in AI and radiomics for bladder, kidney, and prostate cancer: a critical review. Cancers (Basel) 16(4):810
Zhao J, Sun Z, Yu Y, Yuan Z, Lin Y, Tan Y, Duan X, Yao H, Wang Y, Liu J (2023) Radiomic and clinical data integration using machine learning predict the efficacy of anti-PD-1 antibodies-based combinational treatment in advanced breast cancer: a multicentered study. J Immunother Cancer. https://doi.org/10.1136/jitc-2022-006514
Krishna S, Sertic A, Liu Z, Liu Z, Darling GE, Yeung J, Wong R, Chen EX, Kalimuthu S, Allen MJ et al (2023) Combination of clinical, radiomic, and “delta” radiomic features in survival prediction of metastatic gastroesophageal adenocarcinoma. Front Oncol. https://doi.org/10.3389/fonc.2023.892393
Varan M, Azimjonov J, MaÇal B (2023) Enhancing prostate cancer classification by leveraging key radiomics features and using the fine-tuned linear svm algorithm. IEEE Access. https://doi.org/10.1109/ACCESS.2023.3306515
Song H, Yang S, Yu B, Li N, Huang Y, Sun R, Wang B, Nie P, Hou F, Huang C (2023) CT-based deep learning radiomics nomogram for the prediction of pathological grade in bladder cancer: a multicenter study. Cancer Imaging 23(1):89
Li C, Chen H, Zhang B, Fang Y, Sun W, Wu D, Su Z, Shen L, Wei Q (2023) Radiomics signature based on support vector machines for the prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Cancers (Basel) 15(21):5134
Khanfari H, Mehranfar S, Cheki M, Mohammadi Sadr M, Moniri S, Heydarheydari S, Rezaeijo SM (2023) Exploring the efficacy of multi-flavored feature extraction with radiomics and deep features for prostate cancer grading on mpMRI. BMC Med Imaging 23(1):195
Du P, Liu X, Shen L, Wu X, Chen J, Chen L, Cao A, Geng D (2023) Prediction of treatment response in patients with brain metastasis receiving stereotactic radiosurgery based on pre-treatment multimodal MRI radiomics and clinical risk factors: a machine learning model. Front Oncol 13:1114194
Ji J, Zhang T, Zhu L, Yao Y, Mei J, Sun L, Zhang G (2024) Using machine learning to develop preoperative model for lymph node metastasis in patients with bladder urothelial carcinoma. BMC Cancer 24(1):725
Wang W, Sheng R, Liao S, Wu Z, Wang L, Liu C, Yang C, Jiang R (2024) LightGBM is an effective predictive model for postoperative complications in gastric cancer: a study integrating radiomics with ensemble learning. J Imaging Inform Med. https://doi.org/10.1007/s10278-024-01172-0
Vamvakas A, Tsivaka D, Logothetis A, Vassiou K, Tsougos I (2022) Breast cancer classification on multiparametric MRI–increased performance of boosting ensemble methods. Technol Cancer Res Treat 21:15330338221087828
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Ethics declarations
Consent to participate
Every human participant should provide their consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mofid, B., Abdollahi, H., Khajetash, B. et al. Machine learning-based radiomics using magnetic resonance images for prediction of clinical complete response to neoadjuvant chemotherapy in patients with muscle-invasive bladder cancer. Egypt J Radiol Nucl Med 56, 180 (2025). https://doi.org/10.1186/s43055-025-01603-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43055-025-01603-0