- 开放访问
- 发布:
- 2025年10月7日
- 1,,,,2一个 NA1,,,,利利朱
- 2,,,,杰克·罗斯
- 3,,,,Yogini Jani
- 4,,,,5,,,,Enrico Costanza6,,,,
- Maisarah Amran3,,,,
- Zeljko Kraljevic7,,,,
- 8,,,,xi bai1,,,,
- 2,,,,M.M.N.S.迪兰9,,,,
- Jayathri Wijayarathne9,,,,
- Ravi Wickramaratne9,,,,
- Folkert W. Asselbergs1,,,,
- 2,,,,10,,,,理查德·J·杜布森(Richard J.B. Dobson)1,,,,7,,,,
- 8,,,,Wai Keong Wong11一个 NA1和
- …Anoop D. Shah1,,,,2,,,,
- 3
- 一个 NA1一个 BMC医学信息学和决策体积25,文章编号: 365((
2025 )引用本文抽象的背景组织良好的电子健康记录(EHR)对于高质量的患者护理至关重要,但是EHR用户界面对于输入结构化信息可能很麻烦,从而导致大多数信息都是自由文本而不是结构化形式。这使得难以为临床目的检索信息并限制了数据的研究潜力。自然语言处理(NLP)已被认为是提高数据质量和完整性的一种方式,但几乎没有证据表明其有效性。我们试图通过开发一个名为MIADE的模块化,可配置的NLP系统来生成此类证据,该系统旨在与EHR集成。
本文介绍了MIADE的设计和伦敦大学学院医院(UCLH)的部署,并旨在使可能希望在其他地方开发或实施类似系统的人受益。
结果
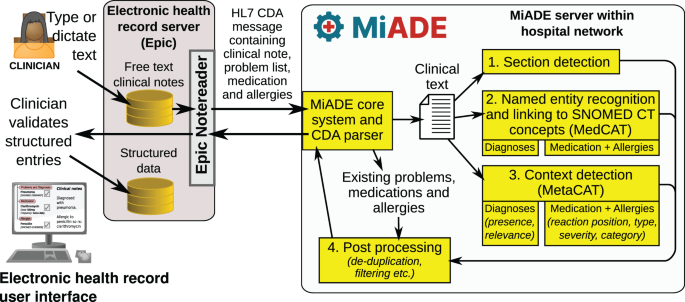
MIADE系统包括从临床注释中提取诊断,药物和过敏的组件,并使用Health 7 Level 7临床文档体系结构(HL7 CDA)消息实时与EHR系统进行通信。
这使得NLP结果可以显示给临床医生进行验证,然后再将其保存到患者的记录中。
Miade将Medcat库(NLP工具的Cogstack家族的一部分)用于指定实体识别(NER),并链接到SNOMED CT以及上下文检测。MEDCAT模型接受了UCLH患者笔记的无监督和监督培训,获得了83.2%的精度(95%CI 77.0,88.1),并召回了85.2%(95%CI 79.1,89.8),用于诊断概念的检测。在模拟测试中,我们发现MIADE将临床医生输入结构化问题清单所花费的时间减少了89%。我们已经在实时临床使用中开始了UCLH的MIADE试验,并与UCLH的EPIC EHR集成在一起。
结论
我们已经开发了一个开源的NLP系统,并成功地将其与EHR集成了一家大型医院的实时临床用途。仿真测试表明,我们的系统大大减少了临床医生进入结构化诊断代码所花费的时间。
背景
电子健康记录中结构化信息的好处
电子健康记录(EHR)中的结构性数据在增强患者安全和促进决策过程中起着至关重要的作用。它可以提供重要信息,例如药物过敏和相互作用的警报,并提醒监视慢性病[1]。结构化信息对于研究和次要数据使用也至关重要[2]。一项随机对照试验表明,结构化问题列表导致临床决策更好,更快[3]。
但是,当今记录中的许多信息都是以自由文本为单位的,而不是结构化的形式[4,,,,5],很难在临床上或研究。例如,在COVID-19大流行期间对UCLH进行的审核发现,问题清单中只有62%的住院诊断[4]。由专业记录标准机构(PRSB)的国家指导建议诊断和其他关键信息的结构化记录和其他关键信息。6]。广泛使用受控临床术语,例如Snomed CT(医学临床术语的系统化命名法)[7]使临床概念能够以一致的方式记录,但前提是临床医生可以轻松使用该系统。对于临床医生来说,在许多EHR系统中输入详细的结构化信息可能很繁重,而且很耗时[5],并且在数据输入上花费的时间会影响人类临床咨询的经验[8,,,,9]。
自然语言处理在护理点的作用
自然语言处理(NLP)可用于将非结构化文本转换为结构化数据[10],但是人类语言的语义细微差别意味着此过程容易出错,并且需要临床医生验证,然后才能安全地将信息用于临床决策。
但是,与有关医疗保健文本回顾性NLP的广泛文献相比,在护理点上有关于NLP的稀疏研究[10,,,,11,,,,12]。Havrilla等。在儿科学术医学中心中实施了针对人类表型本体(HPO)术语进入人类表型本体(HPO)的申请,该术语将数据输入时间从每位患者的15分钟减少到5分钟[13]。Brigham and妇女医院开发了一种与Epic Newereader集成过敏的NLP系统[14]。Solventum销售的商业系统用于护理点的CT编码[15],细微差别[16]和其他人,但有限的发布数据的数据有限。也未知如何最好地配置此类系统以确定结构化信息的最重要项目,并避免无关紧要或不重要的结果的混乱。
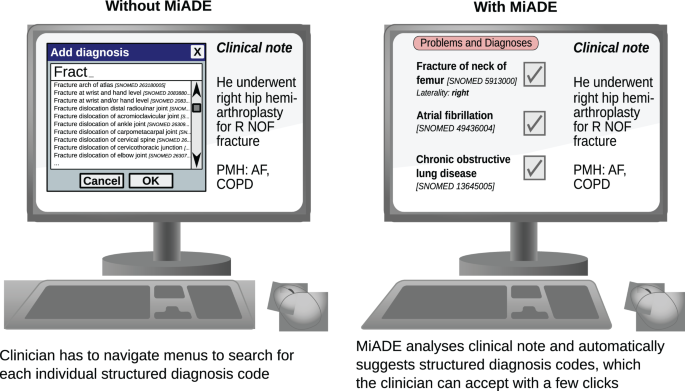
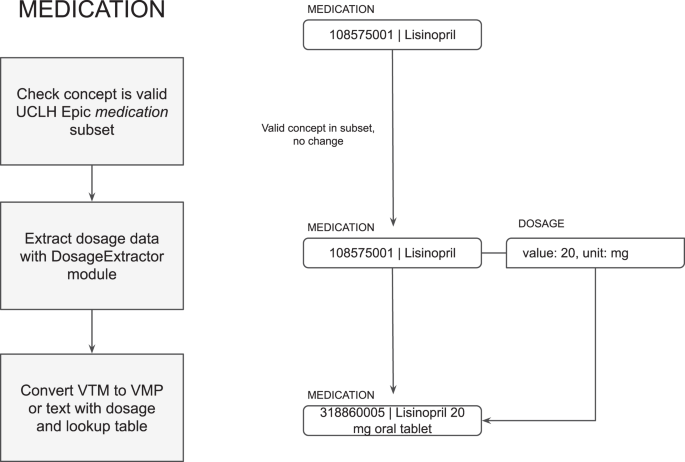
我们的研究旨在通过开发,实施和评估称为MIADE(医学信息AI数据提取器)的新型护理NLP系统来解决这一证据差距。MIADE为临床医生提供了一种替代方法,可以自动为临床医生生成建议以验证的建议,而不是必须浏览Snomed CT词典以找到适当的概念(见图,见图。1)。假设是MIADE将通过使临床医生更容易,更快地输入数据来提高结构化数据的完整性。希望这将改善临床决策和患者的安全,并有益于患者数据(例如研究)的下游用途。
执行
我们开发了一个模块化的开源NLP系统,称为MIADE,能够与EHR和处理临床笔记进行通信,并将建议返回EHR,以展示给临床医生。临床医生可以使用EHR用户界面接受或拒绝MIADE建议,并将其与现有信息调和。
MIADE使用开源Medcat程序[17],NLP工具的Cogstack家族的一部分[18],作为其命名实体识别(NER)工具。MIADE没有自己的用户界面,但使用数据标准健康级别7临床文档体系结构(HL7 CDA)消息与EHR进行通信,并且可以很容易地适应使用快速医疗保健互操作可互操作资源(FHIR)。在UCLH,用户界面由Epic EHR的NoteReader组成部分组成。
系统架构如图所示。2,这说明了系统的密钥函数如何链接在一起:
-
(1)
部分检测
-
(2)
命名实体识别并链接到Snomed CT概念(MEDCAT)
-
(3)
上下文检测(metacat)
-
(4)
后处理
部分检测
虽然临床文件的散文差异很大,但我们发现临床医生经常在带有有意义的标头的部分中以列表式格式进行诊断,问题或药物。与散文相比,此类部分内的信息上下文要容易得多。因此,我们设计了一个文本清洁和段落检测过程,该过程可以根据一组可配置的正则表达式(REGEX)来检测截面标题。然后根据段落类型对注释的输出进行后处理,并且仅返回相关的概念。
命名实体识别并链接到Snomed CT概念(MEDCAT)
MIADE使用开源Medcat [17]库为其核心算法。Medcat是一种自我监督的机器学习算法,用于使用任何概念词汇提取概念,可作为开源Python软件包。与Semehr相比19],Bio-Yodie [20],ctakes [21]和metamap [22]。在大型文本语料库中对Medcat模型进行了无监督的训练,以了解围绕概念的上下文提及以进行歧义。可以进行补充监督培训,以帮助Medcat学习其他同义词以及上下文的正面和负面示例。我们与Medcat开发人员进行了积极的合作,UCLH已经部署了Medcat进行回顾性临床研究[18]。
Medcat概念数据库和培训
我们使用不同的概念数据库(CDB)建立了和训练的单独模型,以解决问题和药物 /过敏,以允许独立开发和实施每种算法。我们使用了一组R脚本和rdiagnosislist r软件包来处理Snomed CT词典并准备概念数据库[23],使用英国Snomed CT 2022年5月版本。脚本和数据可在miade-datasets github存储库中获得(https://github.com/uclh-criu/miade-datasets/)。
问题概念数据库包括社会历史的症状,诊断和重要方面(例如,护理需求和住房),但不与家族史概念,过敏概念,正常发现或与问题清单无关的高级概念,例如与疾病相关的状态。在Snomed CT概念描述中表达的首字母缩写词,并从手动策划的列表中添加了少量常见的同义词。
药物和过敏概念数据库包括物质,可能是过敏反应的症状以及药物和设备词典(DM+D)的药物[24]是虚拟治疗部分(VTM)或虚拟药物(VMP)。
Medcat模型接受了无监督和有监督的步骤的培训[17]。无监督的训练步骤采用了自由文本文档和CDB的语料库,并了解了每个概念上下文的矢量嵌入表示。当检测到模棱两可的项时,该上下文表示用于区分概念。在此步骤中,我们从2019年4月开始使用800,000个UCLH临床笔记(主要是进度票据和放电摘要)的语料库(这是在UCLH安装Epic EHR的时候)。
监督的培训过程涉及手动注释“教授Medcat”,以避免不正确的注释(即,与其链接的概念没有相同含义的单词),并学习其他正确的同义词和背景。可以对Medcat进行培训,以提高其在特定感兴趣领域的性能。
由于临床优先级改善医院和初级保健之间的切换,并改善临床编码和报销,因此我们选择了排放摘要作为该项目的最初重点。因此,我们从前面描述的UCLH语料库中提取了400个排放摘要的随机子集。这些文档使用Cogstack提供的MedCattrainer工具[25],遵循一组注释指南(请参阅补充文本)。我们提取了50个排放摘要的单独随机子集进行测试,该摘要接受了双重独立注释。在分歧的情况下,注释是由第三个注释者手动调和的。
上下文检测(metacat)
MEDCAT包括训练小型双向长期记忆(BI-LSTM)神经网络模型的能力,以检测识别的概念背景。这些被称为Metacat模型,可以使用与MedCat模型的监督训练步骤相同的注释数据进行训练。Metacat模型通过标准的神经网络训练程序进行了训练,并以精确,召回和F1分数进行评估。
我们培训了元数据模型在场,,,,关联和横向性问题。对于药物和过敏反应点,无论物质是过敏原还是正在服用的药物sectancecategory,不良反应风险是过敏还是不宽容过敏反应, 和严重程度过敏(请参阅表1)。我们以感兴趣的概念(问题,药物,物质或反应)代替了通用占位符令牌的培训,以防止模型从概念本身中错误地推断上下文(例如,以避免假设概念被怀疑,因为它经常出现在培训数据中)。
将元数据模型的输出提取到元宣言上课并作为主要概念的属性返回。然后,MIADE将一组逻辑规则与截面类型结合使用,以决定概念的最终上下文。例如,如果在以前的病史中发现了一个概念为“无关紧要的”,则该部分的标题优先于元批准。没有截面标题的文本作为散文段落,MIADE可以根据配置设置在散文段落中返回或忽略概念bustical_list_limit。该参数限制了可以从散文返回的概念的数量。在UCLH的实施中,根据临床医生的反馈,避免返回太多无关紧要的概念是可取的。
合成增强的训练元数据模型的数据
我们在培训元数据模型中遇到的一个挑战是注释数据集中的类不平衡的类别:疑似,历史和无关的注释频率远低于确认和现在的频率。为了解决这个问题,我们使用特殊策划的常见短语模式创建了其他合成训练数据,以表达否定,可疑条件,历史条件和过敏,以增加注释的数据集。
为了促进培训过程的透明度和可重复性,并使用合成的增强数据集(一个)简化创建仪表板是为了管理数据集和模型培训过程。这提供了交互式审查和修改培训数据集所需的注释数据和合成数据的数量的能力。它还可以自动平衡班级权重,版本训练参数,并快速可视化与测试集的混淆矩阵。仪表板已包含在Miade包中。
后处理
核心MIADE管道包含令人敬畏的处理器,,,,注释者, 和dosageExtractor模块。管道接收笔记对象并返回列表概念。在清洁文本和检测段落部分标题的功能中提供了笔记目的。令人敬畏的处理器充当主要的应用程序编程接口(API),实现者可以将MEDCAT模型添加到该接口.add_annotator()方法。这注释者类包装单个Medcat模型,并使用我们开发的自定义后处理算法处理模型的输出。用于用药物概念使用dosageExtractor还提供了模块,该模块提取药物剂量并将输出转换为与CDA兼容的格式。MIADE对返回的所有概念进行了删除。
MIADE处理管道是开源的,可与不同的MEDCAT模型一起使用。要初始化MIADE,实施者需要提供MEDCAT模型目录,该目录训练以提取不同类型(问题,药物和过敏)的概念,并在本地文本数据集中进行微调。后处理组件可通过以下所述转换步骤以及用于提取截面标题的以下的转换步骤的一组查找词典配置。也可以将过滤器列表指定为黑名单不相关的概念,可以对其进行修改,而无需重新训练MEDCAT模型。我们的MIADE研究中使用的概念数据库和查找可在公共中获得MIAD-DATASETS的GitHub存储库[26]。
根据需要,可以在配置文件中禁用MIADE管道组件。对于不需要某些后处理步骤(例如剂量提取或药物转化)的用例。因此,实施者能够使用MEDCAT模型创建插件处理管道。
问题后处理
被否定的概念,可疑的概念,历史悠久的环境通过查找词典转换为相应的Snomed CT术语,例如发烧用否定的背景转换为概念呼吸症。没有转换匹配和无关的概念的概念被滤除。我们的实施将历史程序转换为概念的历史(例如冠状动脉搭桥术的历史),但使历史诊断不变。可以通过自定义查找表轻松配置此行为。
药物后处理
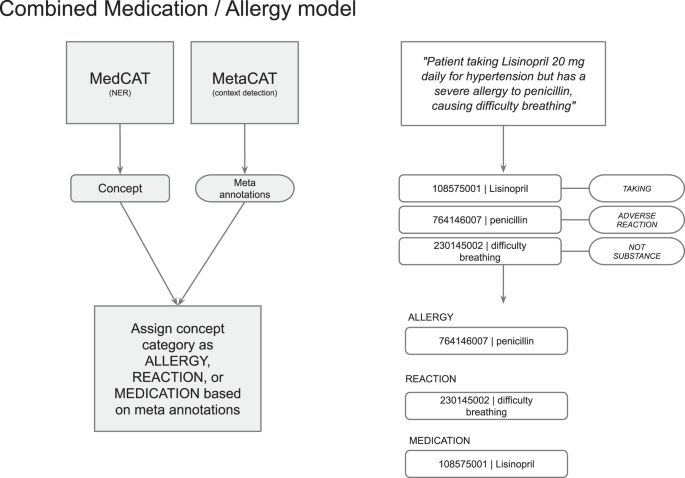
默认的MIAD算法从单个MEDCAT模型中提取药物和过敏,然后根据元批准结果区分并分配药物和过敏类型标签。
首先,检测到的概念是根据基于荟萃注释的药物(服用),过敏(不良反应)或不是物质(不是物质的)处理的。然后,如果适当的话,对该概念进行验证,并将其转换为每个类别中的Snomed CT子集(请参见图。3)。可以根据不同的实现和用例进行配置。
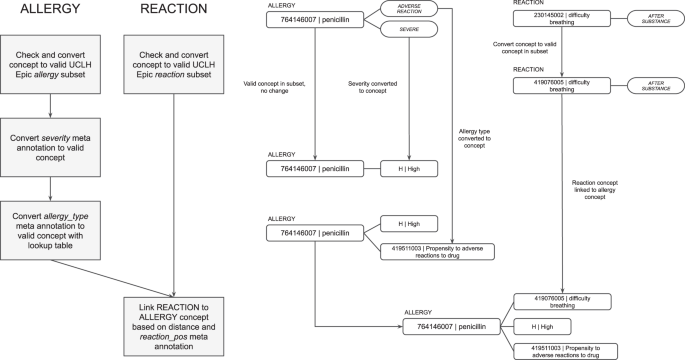
然后,MIADE将过敏原连接到过敏类型,严重性和反应。单个过敏条目作为过敏原CT概念返回,该概念链接到发送给EHR的CDA中的过敏类型,严重性和反应的概念(见图。4)。对于检测到的每种过敏原物质,通过Metacat模型提取严重性并将其转换为适当的SNOMED CT代码。不良反应风险的类型还通过元数据模型提取为“不宽容”,“过敏”或未指定的。根据不良反应风险类型和物质类型选择了过敏记录类型(例如食物不耐受)的总体SNOM CT概念(例如食物不耐症)。最后,根据核心MEDCAT模型检测到该反应,并根据反应概念的荟萃注释是在物质之前还是实质性后的。
如果陈述了剂量形式,则检测到药物为虚拟药物(VMP),或者如果仅陈述该物质,则将药物视为虚拟药物(VTM)。使用dosageExtractor下一节中描述的算法。根据EHR药物词典的设置,VTM可能无效返回EHR,因此MIADE包括一个转换表,将其转换为VMP,以获取常见剂量的口服片剂药物。如果不可能进行转换,则将其作为文本返回,在这种情况下,临床医生在调解NLP输出时需要手动选择适当的VMP。剂量作为与CDA兼容的剂量信息一起返回(请参见图。5)。
提取药物剂量
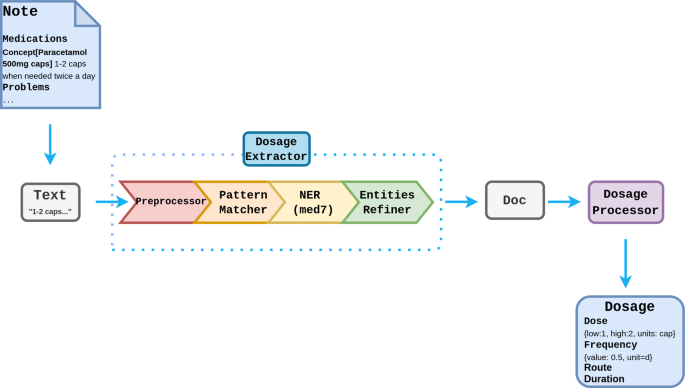
剂量提取的第一阶段是识别构成剂量组成部分的单词和短语。这是使用Med7 ner模型进行的Spacy基于神经网络的处方组件的命名实体识别系统,对模仿III进行培训[27]。基于规则的算法是根据Caliberdrugdose R软件包开发的[28]计算结构化药物剂量。剂量提取器是作为一个Spacy管道可以通过主MIADE API进行配置或独立使用(见图6)。
预处理
预处理步骤是根据caliberdrugdose算法进行的[28],涉及使用两个查找词典(单词和多字)标准化文本中的单词和表达式并纠正常见的拼写错误。其次是数字标准化文本中数量的标准化的步骤(例如,2±200毫克变为400毫克,1/2变为0.5)。
模式匹配
模式匹配器步骤匹配结构性句子模式,并执行基于规则的实体标记,以删除文本中的剂量信息,尤其是遵循特定格式的剂量指令,例如每天两次服用1片,30片。然后,它使用caliberdrugdose的图案查找表来提取剂量结果,并将结果作为属性存储在Spacy Doc目的。
命名实体识别
Med7模型用于提取标签:剂量,药物,持续时间,形式,频率,路线,强度。由于并不需要Med7提取的所有标签,因此对NER结果进行了一些基于规则的改进:
-
将具有相同标签的连续标签组合在一起
-
药物和强度标签被忽略(主要算法被检测为VTM / VMP概念的一部分)
-
强度标签与剂量标签相结合,如果连续
-
如果药物标签遵循剂量(药物应以概念名称,不应以剂量字符串)进行重新标记形式)
CDA剂量类的组成
处理器从查找和NER结果dosageExtractor并将它们解析为兼容CDA剂量班级:
-
剂量[数量,单位,低,高]
-
频率[值,单位,低,高,标准_DEVIATION,INSTICATION_SPECIED,PROPONDITION_ASEREQUIRED]
-
持续时间[值,单位,低,高]
-
路由[代码,displayName]
测试
在将MIADE部署到UCLH之前,我们对MIADE的以下方面进行了测试。首先,我们测试了MEDCAT NLP模型检测SNOMED CT诊断概念的准确性,以确保模型训练足够。其次,我们对与EHR集成的完整管道进行了用户接受测试,以确保临床医生可用并按照预期的方式进行行为。第三,为了验证该系统可以节省临床医生的时间,我们比较了进入有和没有MIADE的问题清单所花费的时间。
最初由于EHR集成问题而无法部署药物和过敏,因此尚未为此功能执行模型测试。需要额外的工作来验证药物和过敏管道准备临床使用。
问题的性能Medcat模型
我们通过在一系列临床专业中测试了50个随机选择的排放摘要笔记的金标准测试集,评估了MIAD MEDCAT模型的问题(症状和诊断)的概念检测的性能(症状和诊断)。这些笔记由健康信息家进行了两倍注释,然后手动审查以解决任何差异。
我们评估了三个组件的MIADE问题算法:概念检测,上下文检测和完整的管道性能。概念检测是指MIADE的NER性能,评估检测到的概念数量,而与从上下文检测算法返回的元数据无关。问题的上下文检测确定肯定状态(确认,否定或怀疑),时间性和相关性。完整的管道性能评估了MIADE是否正确提出了概念检测的完整管道,并根据上下文检测从元数据中应用后处理和过滤逻辑。对于每个组件,我们都以精确,召回和F-1分数量化了性能。对于所有评估,我们都禁用了段落检测和重复数据删除组件,以评估Medcat / Miade算法在解释散文方面的性能。对每个组件进行了两个概念的过滤子集评估 - 症状+诊断(具有SNOMED CT语义类型的概念,发现或疾病,省略了行政概念),并且仅诊断为诊断(主要是SNOMED CT的CT障碍概念,其中一些临床上重要的发现,包括诸如seizure ceizure ceizure)。
概念检测和完整的管道性能以人均为基础评估,而没有概念的位置匹配。由MIADE检测到但在黄金标准中未存在的一个sn子CT概念被归类为假阳性,反之亦然。
为了评估上下文检测,我们需要将MIADE输出中的概念与注释的黄金标准中的概念相匹配。由于概念的变化,我们无法直接执行此操作,从而从文本清洁和预处理阶段进行。相反,对于每个文档,我们首先将MIADE返回的一组概念与黄金标准概念集进行了交叉加入。然后,我们根据位置匹配和字符串相似性的组合使用了一种自定义模糊匹配算法来识别与相同概念相对应的跨度。从此组中,我们检查了每个匹配概念的snomed ID是否匹配,并给出了最终的一组正确检测和建议的概念。上下文检测评估不包括被识别为假阳性,假否定的概念,或者与模糊匹配算法不匹配。
用户接受测试
MIADE安装在链接到EPIC测试安装的服务器上。从事MIADE的临床医生研究人员通过模拟患者历史尝试了该系统,以确保用户界面按预期工作。这是必不可少的,因为史诗般的CDA数据经常缺乏全面文档,因此需要进行反复试验,以验证信息是否按预期显示。
数据输入的测试速度
我们为临床医生设置了一个数据输入任务,并使用输入SNOMED CT概念的默认EHR方法(使用EHRS术语浏览器)和使用MIADE(在此问题将自动进行临床医生验证;请参见图。1)。要求临床医生在一组20个模拟临床笔记上执行任务,每个临床注释包含一个问题清单,其中有10个问题。问题名称是从2019年在UCLH问题列表中随机选择诊断概念的Snomed CT同义词,以随机选择。此任务旨在确定使用MIADE的最大可能节省的时间,因为这些术语是没有变化或错误拼写的术语CT名称,并将由MIADE准确检测到。
研究参与者是五位临床医生,他们有使用EPIC的经验,具有一系列资历和对数字技术的熟悉程度。
与EHR集成
我们将MIADE部署并集成到UCLH的Epic EHR系统中。除了核心开源MIADE库外,我们还开发了一台服务器和CDA解析器,以集成到EHRS中。The MiADE server is deployed as a Docker container on hospital internal network and communicates with Epic over secure HTTPS.It implements the Simple Object Access Protocol (SOAP) methodProcessDocument, which is invoked by Epic to send over a CDA document that contains the note entered by the user in Epic NoteReader.To trigger this request, the user selects or toggles the “Send to NoteReader†button when they are in the Epic NoteReader interface.The input CDA is parsed and processed by the MiADE server, and a response CDA document containing the extracted data is returned to Epic.
We implemented the SOAP endpoint invoked by Epic as a Web Server Gateway Interface (WSGI) sub-application mounted to aFastAPIapp at/notereader/, so that the server is able to receive both RESTful and SOAP requests.This modularisation lends extensibility to the server implementation and allows possibility of integration with different EHR systems, including the option to receive and return data in FHIR format.
Pathway to deployment
MiADE was initially incorporated into a test environment of Epic at UCLH, which is not linked to the live clinical system and does not include actual patient data.Prior to production deployment of MiADE, we load tested our servers using the load testing framework刺槐for a maximum capacity of 100 concurrent users to ensure the server is able to support the load of the feasibility trial.The server was distributed across 8 worker nodes (one for each CPU core), with load balancing handled bynginx。Test request data used were CDA document requests with varying lengths of structured data and progress note lengths.
We developed a clinical safety case following the NHS digital safety standards [29,,,,30]。We performed a risk evaluation by convening hazard workshops with clinicians, health informaticians and software engineers.A hazard log was compiled based on the workshop discussions.Risks fell into three broad themes: risks due to failure of the MiADE software (crashes/data leaks etc.), risks due to the effect of the integration of MiADE on the EHRs (uncovering bugs in an otherwise unused function of the EHRs), and clinician behaviour change (failure to properly vet suggestions from MiADE).
We worked with the EHR specialist team at the hospital to carry out a series of tests to validate that the communication between the EHRs and MiADE was working, data was correctly placed into the medical record, and that the interface functioned as expected.
Following testing and hazard workshops, a safety case was submitted to the Trust Clinical Safety Committee, and subsequent Trust approvals were sought for live deployment within the context of an evaluation study for up to 100 users.The initial live implementation was limited to diagnosis only due to unresolved issues with the Epic EHR integration for medications and allergies.
结果
Performance of the MiADE MedCAT model for problems
Performance of the MiADE MedCAT model for diagnoses was tested on a double-annotated gold standard dataset of 50 discharge summaries containing 551 symptom or diagnosis concepts.
Concept detection - NER
Concept detection refers to pure NER performance by the MedCAT models used in MiADE, with additional filtering for subsets only.Precision was 78.9% (95% Confidence Interval (CI) 74.5, 82.7) and recall was 85.0% (95% CI 80.9, 88.4), giving an F-1 score of 0.82 for ‘Symptoms + Diagnoses’.The number of false positive suggestions was reduced in the ‘Diagnoses Only’ subset with the more restrictive filtering, giving an improved precision of 83.2% (95% CI 77.0, 88.1), a recall of 85.2% (95% CI 79.1, 89.8), and an F-1 score of 0.84;see Table2。Inter-annotator agreement for the correctness of the chosen SNOMED CT concept (Cohen’s kappa) was 0.704.Table 2 Performance of named entity recognition (NER) and full pipeline (NER with context detection) for SNOMED CT symptoms and diagnoses (clinical findings) and diagnoses (disorders) only
The majority of false negatives were due to mis-spellings or variants of phrasing that did not correspond to SNOMED CT synonyms.
Context detection - MetaCAT models
The context detection refers to the combined performance of the MetaCAT models within MiADE and the subsequent post-processing pipeline of filtering and conversion based on the context annotations from MetaCAT.A detected concept is suggested if:
$$\eqalign{& {\rm{Presence}} = \cr & {\rm{Confirmed}} \wedge \cr & ({\rm{Relevance}} = {\rm{Present}} \vee {\rm{Relevance}} = {\rm{Historic}}) \cr} $$
Where ‘Presence’ and ‘Relevance’ refer to annotation outputs from the respective trained MetaCAT models.This was calculated only on the 261 concepts (out of 551) that had been correctly detected in the gold standard dataset and where we were able to match up the concept in the MiADE output with the gold standard using our fuzzy-matching algorithm.
For the ‘Symptoms + Diagnoses’ subset, a total of 228 correctly identified concepts by MiADE were matched to the gold standard dataset, giving a precision of 82.5% (95% CI 76.7, 86.9), a recall of 98.9% (95% CI 95.9, 99.7), and an F-1 score of 0.9.For the ‘Diagnoses Only’ subset, a total of 121 concepts were matched to the gold standard, giving a precision of 80.7% (95% CI 72.7, 86.8).As there were no false negatives from the more restrictive filtering, the recall was 100% (95% CI 96.2, 100) and the F-1 score was 0.89 (Table3)。
Full pipeline
The full pipeline performance of MiADE includes the concept detection (NER), context detection (MetaCAT models), post-processing pipeline of filtering and conversion based on the context detection, and final filtering to ‘Symptoms + Diagnoses’ and ‘Diagnoses Only’ subsets of concepts.
The precision of the ‘Symptoms + Diagnoses’ subset was 60.7% (95% CI 54.5, 66.5) and the recall was 74.3% (95% CI 68, 79.7) with an F-1 score of 0.67.Like the NER results, the full pipeline ‘Diagnoses Only’ subset had fewer false positives, giving a precision of 64.8% (95% CI 55.9, 72.7), a recall of 75.2% (95% CI 66.2, 82.5) and an F-1 score of 0.7 (Table2)。Testing the speed of data entry
Five doctors - two Specialty Registrars, an Internal Medicine Training (IMT) doctor, a Foundation Year 2 (FY2) doctor and a Foundation Year 1 (FY1) doctor - participated in the speed test with simulated clinical notes.
The results showed that MiADE and NoteReader reduced the time taken to enter problem lists by 89% compared to the default Epic interface (see Table4)。
部署
We successfully deployed MiADE integrated in a live EHR environment at UCLH.MiADE was deployed on a microservice architecture within UCLH internal network.This instance of MiADE runs on distributed CPU, but MiADE is also capable of running on GPU or scaled to a cloud-based approach using Kubernetes.
MiADE went live for internal investigators on the project on 14 February 2024 and became available to use for all trial users from 26 February 2024. From the period of 26 February 2024 to 8 May 2024, a total of 1645 documents were processed by MiADE, and 317 concepts were extracted and added to 111 patients’ records.
讨论
Summary of main findings
We successfully developed an open source NLP system called MiADE and integrated it with the EHR at UCLH for an evaluation in live clinical use.MiADE is designed to integrate with EHRs to extract diagnoses, medication and allergies from a clinical note in real time and provide the results for immediate clinical verification.In simulation testing we found that MiADE can significantly reduce the time taken for clinicians to enter structured problem lists.We hope the information in this article will be useful for others proposing similar work in the future.
Algorithm performance and clinical utility
We consider that the performance of the MiADE MedCAT model (83% precision, 85% recall for NER on diagnoses) is sufficient to be clinically useful, and these performance statistics are similar to those obtained using a range of methods on I2b2–2010 concept extraction benchmarks in a recent systematic review [31]。While some diagnoses may be incorrect and some may be missed, these are unlikely to cause safety issues because MiADE operates within a human-in-the-loop system;i.e. the EHR user interface requires manual review of MiADE suggestions (with an opportunity to correct them) before they are committed to the patient’s record.
The detection of context and relevance in prose was more difficult and reduced the F1 score to 0.67, so in production we configured MiADE to return only problems that are in a paragraph or section with a heading such as “Problem list†or “Diagnosesâ€.Clinicians can therefore use headings to lay out their clinical note for MiADE to work efficiently.
Algorithm design
The core MiADE algorithm consists of MedCAT models, which perform NER + linking, and MetaCAT models, which augment concepts detected by the NER step with contextual information from LSTM-powered MetaCAT models.The MiADE system is modular, customisable, and portable to different hospital EHRs needs.It does this by allowing implementers to build plug-and-play pipelines of different MedCAT models and configure custom post-processing components.
The role of small models such as MedCAT has been questioned in the era of LLMs.Whilst LLMs have impressive reasoning ability, they are error-prone for medical coding tasks (e.g. they can hallucinate SNOMED CT concepts that do not exist) [32] and in general do not perform as well as smaller models on NER tasks [33]。They would require additional manual processing to ensure they return reliable results, in which case they would not have any significant advantage over smaller models.In addition, the resource and environmental cost of LLMs make them difficult and expensive to deploy, fine-tune, and scale.However, LLMs are superior at understanding context and can perform complex reasoning tasks [34,,,,35]。This could be used to improve the ability of MiADE to extract concepts from prose, and also to gather and generate more detailed structured information.Recent advances in LLM techniques such as Retrieval Augmented Generation (RAG) [36], chain-of-thought prompting [35] and agentic workflows [37] are promising approaches.
With this in mind, MiADE is designed to be modular and extensible, allowing components like the NER model or post-processing pipelines to be easily replaced with more sophisticated models or algorithms.The plug-and-play architecture of MiADE enables the creation of robust pipelines that combine fast, lightweight algorithms in simple components with LLMs for context-aware reasoning in more complex components.MiADE should serve as a foundation for building open, hybrid-approach data extractor pipelines that can be easily fine-tuned and integrated at different hospital EHRs.We believe this will be a significant advantage over existing monolithic enterprise solutions on the market, such as Solventum [15] and Nuance [16]。By leveraging the power of open source, our approach empowers the community to create customised versions of MiADE tailored to their individual needs.This flexibility and adaptability will set MiADE apart from the rigid, one-size-fits-all commercial offerings currently available.
Challenges encountered
A particular challenge we encountered while developing MiADE for clinical deployment was working with local configurations of EHRs data.In the case of healthcare interoperability standards, function does not always follow form - although HL7 standards technically ensures interoperability between systems, we have found that local preferences in coding systems and configurations may deviate from the out-of-the-box template specified by the EHR provider’s developer resources for third-party integration, leading to inaccurate parsing of data.These variations in usage are often not well documented, and as a result, a disproportionate amount of time was needed to coordinate with different stakeholders and reverse engineer the behaviour of the reading and writing of data in the EHRs test instance.In other words, our experience indicates that the reality of existing EHR systems is that they are interoperable, but notcomposable。This may present more overhead for widespread roll-out and installation of MiADE at different sites, adding to the existing challenges of working with legacy systems.
Future implementers should also consider appropriate strategies for development infrastructure and Trusted Research Environments (TRE) optimised for the clinical deployment of machine learning and NLP workflows [38]。While accessing hospital systems via secure network is adequate for ad hoc development work and server hosting, the overhead and technical debt can quickly add up for work that involves data and model training, taking away from time that could be spent on setting up robust Continuous Integration / Continuous Delivery (CI / CD) software development and MLOps (Machine Learning Operations) best practices.
限制
Although MiADE was developed with close consultation of clinicians and end-users, the user interface design and what can be presented to a user is limited by the fact that outputs have to adhere to the Epic NoteReader UI interface.For example, the MiADE backend has no control over the order in which the problems are displayed (unless explicitly coerced by adding numbering before the concept text), or the action buttons presented to users within the interface.To ensure user-friendliness and more wide-spread clinician adoption of the system, full control over user interface design should be considered.The interfacing method with Epic NoteReader also dictates that CDA must be used for communication between Epic and NLP vendors, which can be more limiting in terms of the flexibility of information communication than FHIR.
While the lightweight MedCAT NER models excel at quickly detecting well-written concepts, they have limited ability to understand the context of concepts, and can have difficulty with ambiguous text.The presence of unusual spellings and abbreviations in clinical text required us to augment the concept database manually, making the system less generalised.The text searching algorithm of the core NER models is vulnerable to bracketing mistakes, for example: “abnormalities found in lung, cancer not detected†might lead MedCAT to detect the concept “lung cancerâ€.The MetaCAT models that perform the context detection could similarly mistake nearby but semantically unrelated context when classifying a concept.
The MiADE post-processing algorithm, in particular medications and allergies, was written tightly-coupled to the peculiarities of how the local hospital instance stores medication concepts.As a result the post-processing algorithm may not be readily applicable to another location without adjustments to the code.We have designed the MiADE pipeline to be extensible to account for this, so that custom logic and data can be created and configured, together with MedCAT models trained on local text.
The MiADE testing reported in this paper has limitations.The NLP performance testing was based on a sample of a single type of clinical note (discharge summaries), and may not be representative of performance in specific clinical areas.The simulation speed test was artificial and may not represent real life performance because MiADE NLP may not work as well with real patient notes.However it was intended to demonstrate the need and theoretical maximal benefit of such a system.
Recommendations for future work
This project takes an initial step towards making EHR user interfaces more intelligent and user-friendly in order to capture richer structured data at the point of care.There are further areas of development in the user interface design itself, such as enabling NLP suggestions to be displayed as the user types or dictates rather than on saving the document, as with NoteReader.Diagnoses are currently recorded as flat SNOMED CT concepts without attributes such as date, body, site, manifestations etc. which are included in openEHR [39] and FHIR [40] information models for diagnoses.More sophisticated NLP techniques such as LLMs can be used to detect complex concepts and their relationships, while maintaining accurate coding.There is also a need to understand how well models trained at one centre perform at another, and to what degree local training of models and tailoring of algorithms is required.
结论
We have successfully designed and implemented an open source NLP system to extract structured information from free text at the point of care, and integrated it with the EHR system in live clinical use at a major hospital.Simulation testing has shown that our system significantly reduces the time taken for clinicians to enter structured diagnosis codes, and it may therefore help to improve the completeness of clinical records without impeding clinicians’ workflow.
Availability and requirements
The MiADE package version v1.0.7 used in this paper is available under the Elastic 2.0 licence.It can be accessed and used through the following means:
-
项目名称: MiADE
-
Project home page: Detailed documentation is available at (https://uclh-criu.github.io/miade/).
-
操作系统: Linux, macOS
-
编程语言: Python
-
执照: Elastic 2.0
-
Any restrictions to use by non-academics: Under the Elastic 2.0 licence, you may not provide the products to others as a managed service
数据可用性
The patient data used for training NLP models may contain sensitive information and external access is therefore not possible.The trained NLP models may be shared with other NHS Trusts according to data sharing agreements.Datasets of SNOMED CT concepts, mappings and lookups used by the UCLH implementation of MiADE are available on GitHub (https://github.com/uclh-criu/miade-datasets/)。
材料可用性
不适用。
代码可用性
The MiADE package version v1.0.7 used in this paper is available under the Elastic 2.0 licence.It can be accessed and used through the following means:
•PyPI (Python Packaging Index): MiADE is available as a Python package on PyPI (https://pypi.org/project/miade/)。It can be installed using pip:
pip install miade= = 1.0.7
•源代码: The latest version is available on GitHub (https://github.com/uclh-criu/miade)。
文档: Detailed installation instructions, usage examples, and API reference, is available at:https://uclh-criu.github.io/miade/
•档案: A static version of the code has been archived and assigned a DOI: [DOI 10.5281/zenodo.13323598]
•要求: MiADE is compatible with Python > = 3.8, < 3.12 and has been tested on Linux and macOS.A complete list of dependencies and version constraints can be found in the pyproject.toml and requirements.txt files in the GitHub repository.
•可重复性: The scripts and data used to train the MedCAT models and post-processing lookup data used in the MiADE implementation at UCLH is made available in the Github repository miade-datasets (https://github.com/uclh-criu/miade-datasets)。
The “Rdiagnosislist†R package was used for processing of SNOMED CT dictionaries and preparation of concept databases for MiADE.The latest version is available on GitHub (https://github.com/anoopshah/Rdiagnosislist), licensed under GPL-3.The stable version is on the Comprehensive R Archive Network (https://cran.r-project.org/web/packages/Rdiagnosislist/index.html)。
缩写
- CDA:
-
Clinical Document Architecture
- CDB:
-
Concept database
- EHR:
-
Electronic health record
- FHIR:
-
Fast Healthcare Interoperable Resources
- HL7:
-
Health Level 7
- HPO:
-
Human Phenotype Ontology
- LLM:
-
大语言模型
- LSTM:
-
Long short term memory
- MedCAT:
-
Medical Concept Annotation Tool
- MiADE:
-
Medical information AI Data Extractor
- NER:
-
Named Entity Recognition
- NHS:
-
National Health Service
- NIHR:
-
National Institute for Health and Care Research
- NLP:
-
自然语言处理
- SNOMED CT:
-
Systematized Nomenclature of Medicine Clinical Terms
- NLP:
-
自然语言处理
- UCLH:
-
University College London Hospitals
- UMLS:
-
Unified Medical Language System
- VMP:
-
Virtual Medicinal Product
- VTM:
-
Virtual Therapeutic Moiety
参考
Weed LL.Medical records that guide and teach.N Engl J Med。1968;278(11):593–600.https://doi.org/10.1056/NEJM196803142781105。CAS
一个 PubMed一个 Google Scholar一个 Dugas M, Blumenstock M, Dittrich T, Eisenmann U, Feder SC, Fritz-Kebede F, Kessler LJ, Klass M, Knaup P, Lehmann CU, Merzweiler A, Niklas C, Pausch TM, Zental N, Ganzinger M. Next-generation study databases require FAIR, EHR-integrated, and scalable electronic data capture for medical documentation and decision support.
NPJ Digit Med。2024;7(1):10.https://doi.org/10.1038/s41746-023-00994-6。PubMed
一个 PubMed Central一个 Google Scholar一个 Klappe ES, Heijmans J, Groen K, Ter Schure J, Cornet R, Keizer NF.Correctly structured problem lists lead to better and faster clinical decision-making in electronic health records compared to non-curated problem lists: a single-blinded crossover randomized controlled trial.
Int J Med Inf。2023;180(105264), 105264 (https://doi.org/10.1016/j.ijmedinf.2023.105264。Poulos J, Zhu L, Shah AD.
Data gaps in electronic health record (ehr) systems: an audit of problem list completeness during the covid-19 pandemic.Int J Multiling Med Inf.2021;150, 104452 (https://doi.org/10.1016/j.ijmedinf.2021.104452。Kalra D, Fernando B. Approaches to enhancing the validity of coded data in electronic medical records.
Prim Care Respir J. 2011;20(1):4–5.https://doi.org/10.4104/pcrj.2010.00078。PubMed
一个 Google Scholar一个 Shah AD, Quinn NJ, Chaudhry A, Sullivan R, Costello J, O’Riordan D, Hoogewerf J, Orton M, Foley L, Feger H, Williams JG.Recording problems and diagnoses in clinical care: developing guidance for healthcare professionals and system designers.
BMJ Health Care inform.2019;26(1).https://doi.org/10.1136/bmjhci-2019-100106。Millar J. The need for a global language - SNOMED CT introduction.
Stud Health Technol inform.2016;225:683–85.
PubMed一个 Google Scholar一个
Pearce C, Dwan K, Arnold M, Phillips C, Trumble S. Doctor, patient and computer–a framework for the new consultation.Int J Med Inf。2009;78(1):32–38.https://doi.org/10.1016/j.ijmedinf.2008.07.002。Google Scholar
一个 Theadom A, Lusignan S, Wilson E, Chan T. Using three-channel video to evaluate the impact of the use of the computer on the patient-centredness of the general practice consultation.
Inf Prim Care.2003;11(3):149–56.https://doi.org/10.14236/jhi.v11i3.563。Google Scholar
一个 Sedlakova J, Daniore P, Horn Wintsch A, Wolf M, Stanikic, Haag C, Sieber C, Schneider G, Staub K, Alois Ettlin D, Grübner O, Rinaldi F, Wyl V. University of Zurich digital society initiative (UZH-DSI) health community: challenges and best practices for digital unstructured data enrichment in health research: a systematic narrative
审查。PLOS数字健康。2023;2(10), 0000347 (https://doi.org/10.1371/journal.pdig.0000347。Wu H, Wang M, Wu J, Francis F, Chang Y-H, Shavick A, Dong H, Poon MTC, Fitzpatrick N, Levine AP, Slater LT, Handy A, Karwath A, Gkoutos GV, Chelala C, Shah AD, Stewart R, Collier N, Alex B, Whiteley W, Sudlow C, Roberts A, Dobson RJB.
A survey on clinical natural language processing in the United Kingdom from 2007 to 2022. NPJ Digit Med.2022;5(1):186.https://doi.org/10.1038/s41746-022-00730-6。PubMed
一个 PubMed Central一个 Google Scholar一个 Velupillai S, Suominen H, Liakata M, Roberts A, Shah AD, Morley K, Osborn D, Hayes J, Stewart R, Downs J, Chapman W, Dutta R. Using clinical natural language processing for health outcomes research: overview and actionable suggestions for future advances.J Biomed Inf.
2018。https://doi.org/10.1016/j.jbi.2018.10.005。Google Scholar
一个 Havrilla JM, Singaravelu A, Driscoll DM, Minkovsky L, Helbig I, Medne L, Wang K, Krantz I, Desai BR.
PheNominal: an EHR-integrated web application for structured deep phenotyping at the point of care.BMC Med。inf。决策。mak。2022;22(Suppl 2):198.https://doi.org/10.1186/s12911-022-01927-1。Google Scholar
一个 Zhou L. AHRQ grant final progress report: encoding and processing patient allergy information in EHRs.
Technical report.2018。https://digital.ahrq.gov/sites/default/files/docs/citation/r01hs022728-zhou-final-report-2018.pdf,Brigham and Women’s Hospital, Harvard Medical School.
3M m*modal fluency direct.https://www.solventum.com/en-us/home/h/f/b5005151015/。Clintegrity Physician coding.
https://www.nuance.com/healthcare/provider-solutions/coding-compliance/physician-coding.html。Kraljevic Z, Searle T, Shek A, Roguski L, Noor K, Bean D, Mascio A, Zhu L, Folarin AA, Roberts A, Bendayan R, Richardson MP, Stewart R, Shah AD, Wong WK, Ibrahim ZM, Teo JT, Dobson RJB.Multi-domain clinical natural language processing with medcat: the medical concept annotation toolkit.
Corr Abs/2010.01165.2020。https://arxiv.org/abs/2010.01165。Noor K, Roguski L, Bai X, Handy A, Klapaukh R, Folarin A, Romao L, Matteson J, Lea N, Zhu L, Asselbergs FW, Wong WK, Shah A, Dobson RJ.
Deployment of a free-text analytics platform at a UK National health Service research hospital: cogStack at University college London hospitals.JMIR Med inform.2022;10(8), 38122 (https://doi.org/10.2196/38122。Wu H, Toti G, Morley KI, Ibrahim ZM, Folarin A, Jackson R, Kartoglu I, Agrawal A, Stringer C, Gale D, Gorrell G, Roberts A, Broadbent M, Stewart R, Dobson RJB.
SemEHR: a general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research.J. Am。医学inf。assoc.2018;25(5):530–37.https://doi.org/10.1093/jamia/ocx160。Google Scholar
一个 Bio-YODIE.
https://github.com/GateNLP/Bio-YODIE。Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG.Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications.
J. Am。医学inf。assoc.2010;17(5):507–13.https://doi.org/10.1136/jamia.2009.001560。Google Scholar
一个 MetaMap.
http://metamap.nlm.nih.gov/。Shah AD: Rdiagnosislist: Manipulate SNOMED CT Diagnosis Lists.R package version 1.5.0.
2025。https://cran.r-project.org/web/packages/Rdiagnosislist/index.html。Business Services Authority N. Dictionary of medicines and devices.
https://dmd-browser.nhsbsa.nhs.uk/。Searle T, Kraljevic Z, Bendayan R, Bean D, Dobson R. MedCATTrainer: a biomedical free text annotation interface with active learning and research use case specific customisation.Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing.
(EMNLP-IJCNLP):, Stroudsburg, PA, USA: System Demonstrations.Association for Computational Linguistics;2019, pp. 139–44).https://www.aclweb.org/anthology/D19-3024。GitHub repository miade-datasets.
https://github.com/uclh-criu/miade-datasets。Kormilitzin A, Vaci N, Liu Q, Nevado-Holgado A. Med7: a transferable clinical natural language processing model for electronic health records.arXiv preprint arXiv:2003.01271 (2020.
Shah AD, Martinez C. An algorithm to derive a numerical daily dose from unstructured text dosage instructions.
药epidemiol药物SAF。2006;15(3):161–66.
PubMed一个 Google Scholar一个
White S. Clinical risk management: Its application in the manufacture of health it systems - specification.Technical report.2018。https://digital.nhs.uk/data-and-information/information-standards/information-standards-and-data-collections-including-extractions/publications-and-notifications/standards-and-collections/dcb0129-clinical-risk-management-its-application-in-the-manufacture-of-health-it-systems,NHS Digital.
White S. Clinical risk management: Its application in the deployment and use of health it systems - specification.Technical report.2018。https://digital.nhs.uk/data-and-information/information-standards/information-standards-and-data-collections-including-extractions/publications-and-notifications/standards-and-collections/dcb0160-clinical-risk-management-its-application-in-the-deployment-and-use-of-health-it-systems,NHS Digital.
Fraile Navarro D, Ijaz K, Rezazadegan D, Rahimi-Ardabili H, Dras M, Coiera E, Berkovsky S. Clinical named entity recognition and relation extraction using natural language processing of medical free text: a systematic review.Int J Med Inf。2023;177(105122), 105122 (https://doi.org/10.1016/j.ijmedinf.2023.105122。Soroush A, Glicksberg BS, Zimlichman E, Barash Y, Freeman R, Charney AW, Nadkarni GN, Klang E. Large language models are poor medical coders — benchmarking of medical code querying.
In: NEJM AI.卷。1(5), 2300040 (Publisher: Massachusetts Medical Society; 2024.https://doi.org/10.1056/AIdbp2300040。Accessed 2024-08-09.
Zhao WX, Zhou K, Li J, Tang T, Wang X, Hou Y, Min Y, Zhang B, Zhang J, Dong Z, Du Y, Yang C, Chen Y, Chen Z, Jiang J, Ren R, Li Y, Tang X, Liu Z, Liu P, Nie J-Y, Wen J-R.A survey of large language models.2023。https://arxiv.org/abs/2303.18223。Kojima T, Gu SS, Reid M, Matsuo Y, Iwasawa Y. Large language models are Zero-Shot Reasoners.
2023。https://arxiv.org/abs/2205.11916。Wei J, Wang X, Schuurmans D, Bosma M, Ichter B, Xia F, Chi E, Le Q, Zhou D. Chain-of-thought prompting elicits reasoning in large language models.
2023。https://arxiv.org/abs/2201.11903。Xiong G, Jin Q, Lu Z, Zhang A. Benchmarking retrieval-augmented generation for medicine.
2024。https://arxiv.org/abs/2402.13178。Sudarshan M, Shih S, Yee E, Yang A, Zou J, Chen C, Zhou Q, Chen L, Singhal C, Shih G. Agentic LLM workflows for generating patient-friendly medical reports.
2024。https://arxiv.org/abs/2408.01112。Harris S, Bonnici T, Keen T, Lilaonitkul W, White MJ, Swanepoel N. Clinical deployment environments: five pillars of translational machine learning for health.
2022;4.
Garde S. Ocean health Systems: OpenEHR Problem/Diagnosis archetype.https://ckm.openehr.org/ckm/archetypes/1013.1.169。Accessed: 2021-5-10.https://ckm.openehr.org/ckm/archetypes/1013.1.169。Condition - FHIR v4.0.1.
https://www.hl7.org/fhir/condition.html。Accessed: 2021-2-23.https://www.hl7.org/fhir/condition.html。下载参考致谢This study uses patient information from NHS patients collected as part of their care and support.
The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
资金
This study was funded by the National Institute of Health Research Artificial Intelligence in Health and Care Award (AI_AWARD01864), the Engineering and Physical Sciences Research Council (EP/Y018087/1), and supported by the National Institute for Health and Care Research University College London Hospitals Biomedical Research Centre.
Cogstack was funded by the National Institute of Health Research Artificial Intelligence in Health and Care Award (AI_AWARD02361).ADS was supported by UKRI (Horizon Europe Guarantee for DataTools4Heart) and British Heart Foundation Accelerator Award (AA/18/6/24223).YJ is co-lead for Safer Evidence theme and Equality, Diversity & Inclusion lead for the NIHR Central London Patient Safety Research Collaboration (NIHR204297).The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
道德声明
道德批准并同意参加
The development of MiADE was undertaken as part of Cogstack service development within University College London Hospitals NHS Trust and has approval of relevant Trust committees.The MiADE evaluation study was approved by the Hampshire ethics committee (23/SC/0221: Amendment 1, March 2024) and is registered on ISRCTN (https://www.isrctn.com/ISRCTN58300671)。All clinicians participating in the study gave written informed consent.The study was conducted in accordance with the ethical principles of the Declaration of Helsinki.
同意出版
不适用。
同意出版
作者没有宣称没有竞争利益。
附加信息
Publisher’s Note
关于已发表的地图和机构隶属关系中的管辖权主张,Springer自然仍然是中立的。
电子补充材料
Below is the link to the electronic supplementary material.
权利和权限
开放访问This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.要查看此许可证的副本,请访问http://creativecommons.org/licenses/4.0/。Reprints and permissions
引用本文
Jiang-Kells, J., Brandreth, J., Zhu, L.
等。Design and implementation of a natural language processing system at the point of care: MiADE (medical information AI data extractor).BMC Med Inform Decis Mak25 , 365 (2025).https://doi.org/10.1186/s12911-025-03195-1
已收到:
公认:
出版:
doi:https://doi.org/10.1186/s12911-025-03195-1